The Importance Of Epigenetic Alterations In The Development Of Epstein-Barr Virus-Related Lymphomas
Maria Takacs1, Judit Segesdi2, Ferenc Banati2, Anita Koroknai2, Hans Wolf3, Hans Helmut Niller3 and Janos Minarovits2
1Division of Virology, National Center for Epidemiology, H-1097 Budapest, Gyali út 2-6, Hungary
2Microbiological Research Group, National Center for Epidemiology, H-1529 Budapest, Pihenö u. 1. Hungary
3Institute for Medical Microbiology and Hygiene, University of Regensburg, Franz-Josef-Strauss-Allee 11, D-93053 Regensburg, Germany
Published: November 15 2009
Received: October 10 2009
Accepted: November 13 2009
Medit J Hemat Infect Dis 2009, 1(2): e2009012 DOI 10.4084/MJHID.2009.012
This article is available from: http://www.mjhid.org/article/view/5080
Introduction:
Epstein-Barr virus (EBV), a member of the Gammaherpesvirinae
subfamily, is a herpesvirus widespread in human populations [1]. EBV
infects B lymphocytes via the complement receptor CD21, and the
B-cell-bound virions can be transferred efficiently to CD21-negative
epithelial cells [2]. Thus, when transmitted orally,
EBV enters the
epithelial cells of the oropharynx resulting in productive (lytic) infection.
On the contrary, the EBV-B cell interaction is usually nonproductive,
the latent viral genomes persist as circular episomes and co-replicate
with the cellular DNA once per cell cycle.
The association of latent EBV genomes with malignant tumors is well
documented. The virus has been discovered in cells derived from endemic
Burkitt’s lymphoma samples [3]. In addition, other
lymphomas (X-linked
lymphoproliferative syndrome, posttransplant lymphoproliferative
disease, AIDS-associated lymphoma, peripheral T/NK-cell lymphoma,
Hodgkin’s lymphoma), certain carcinomas (nasopharyngeal carcinoma,
gastric carcinoma, carcinomas of the major salivary glands, thymic
carcinoma, mammary carcinoma) and a sarcoma (leiomyosarcoma) also
regularly carry latent EBV genomes [1,4]
(Table 1).
Sequencing of the 172 kb double stranded
linear genome of the prototype EBV strain (B95-8) significantly
contributed to the identification of latent, growth-transformation
associated proteins of the virus [5].
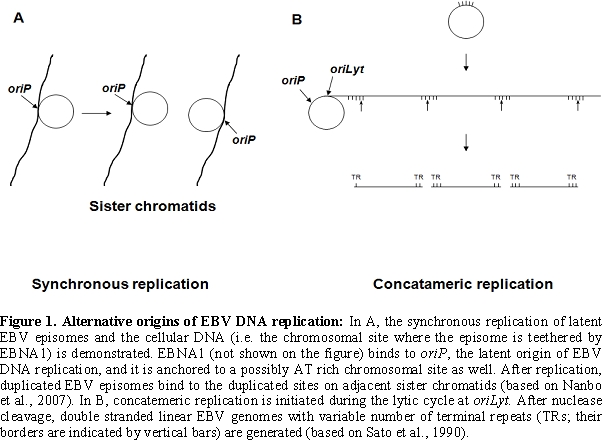
During productive infection all EBV encoded proteins are expressed and
concatemeric replication of circular viral DNA templates is initiated
at oriLyt (the lytic origin
of replication) by the viral DNA polymerase with the help of EBV
encoded transactivator and replication proteins [6,7,8] (Figure
1)
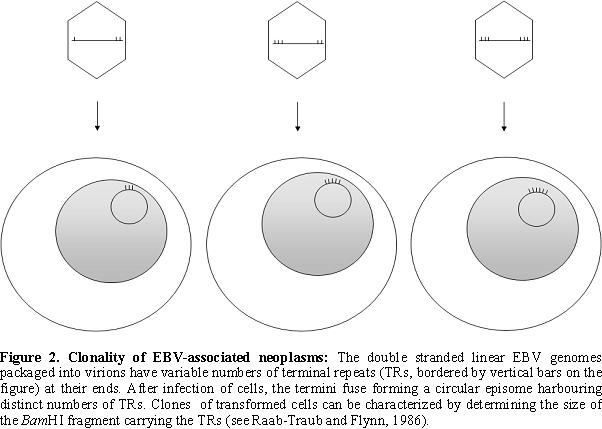
The viral DNA packaged into the virion ends in terminal repetitions (TRs). Soon after in vitro infection of B cells the TRs fuse with each other and circular episomes are generated by amplification of a single copy of the viral DNA. Only a subset of the viral genes are transcribed from the latent EBV episomes that co-replicate with the cellular genome in the emerging B cell clones. It is noteworthy that during latent, non-productive infection, DNA synthesis is initiated at a distinct origin of replication called oriP (that is different from oriLyt), by the cellular DNA replication machinery [9,10] (Figure 1). Expression of growth-transformation-associated viral latency genes leads to the establishment of permanently growing (immortal) lymphoblastoid cell lines (LCLs). The major EBV-associated malignancies also carry predominantly circular EBV DNA molecules. EBV infection is apparently important in their development, because analysis of fused TR sequences showed that these tumors are clonal proliferations of cells infected by EBV on a single occasion, presumably in an early stage of tumorigenesis [11] (Figure 2).
Host
cell phenotype-dependent expression of latent EBV genes: EBV latency
types
The majority of EBV encoded proteins is involved in productive EBV infection
and includes transactivators, enzymes necessary for viral DNA
amplification and structural components of the virions. There is a
sequential order of gene activation in the course of the lytic cycle:
immediate early genes (BZLF1 and BRLF1) are expressed first and
activate the so called early genes. Late proteins are expressed after
viral DNA synthesis [12].
A more restricted pattern of viral gene expression can be observed in latently infected
cells. The latent EBV genomes carried by in vitro transformed LCLs and
lymphoproliferations developing in recipients of organ or bone marrow
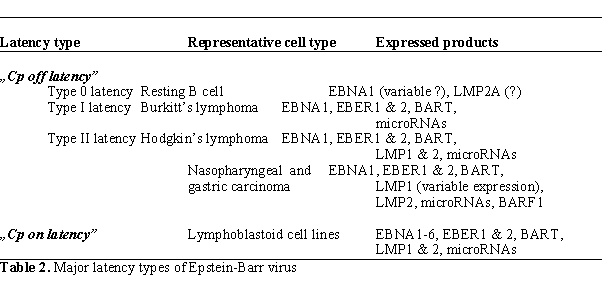
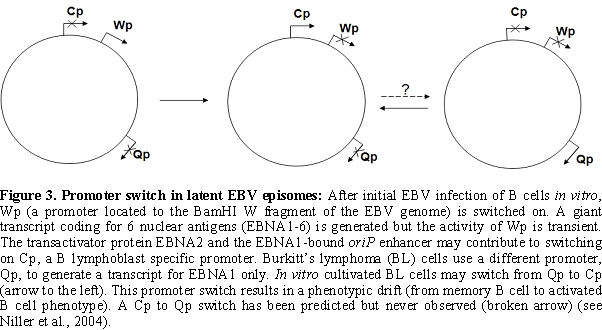
transplants express six nuclear proteins (EBNAs). The EBNA1-6
transcripts are initiated at the C promoter (Cp, located to the BamHI C fragment of the viral
genome), hence the term „Cp on”
latency (also called
latency type III)13 (Table 2, Figure 3).
Cp is active only in B lymphoblasts [14].
EBNA1 is required for stable maintenance of viral episomes, whereas
EBNA2, the major transactivator protein of EBV transactivates both
viral promoters (Cp, LMP1p, LMP2Ap, LMP2Bp, see below) and promoters of
cellular genes (CD23, CD21). EBNA3-6 modulate the transactivator
activity of EBNA2 (see below).
In „Cp on” latency (reviewed
by Niller et al.1) one can also detect three transcripts for latent
membrane proteins (LMP1, LMP2A, LMP2B), two small RNA molecules (EBER 1
and 2), transcripts derived from the BamHI A fragment of the genome
(BARTs, reviewed by Smith15) and a series of microRNAs [16,17,18]. The
transmembrane proteins LMP1 and LMP2 affect various signal transduction
pathways. Expression of LMP1 in rodent cells causes malignant
transformation. The EBERs and BARTs (and the protein products of
BARTs), and the BARF1 protein were also implicated in oncogenesis
(reviewed by Niller et al.1). The microRNAs derived from transcripts of
the BHRF1 gene and the BART transcripts may target both viral and
cellular RNAs affecting thereby the abundance of lytic (BALF5) and
latent (LMP1) EBV transcripts as well as certain cellular RNAs (e.g.
that of CXCL-11, coding for a chemokine) (for review see Swaminathan [19]
).
In other EBV-carrying cell types Cp is switched off („Cp off” latency).
These include memory B cells, Burkitt’s lymphomas (BLs) and BL cell
lines that maintain the BL biopsy phenotype (type I BL cell lines),
EBV-associated carcinomas (nasopharyngeal carcinoma, gastric carcinoma)
and Hodgkin’s lymphomas. In „Cp off
latency”
typically an alternative promoter (Q promoter or Qp) is used for
expression of EBNA1 transcripts, but the transcripts coding for the
other five EBNAs are absent. The various cell types carrying silent C
promoters may differ from each other regarding the expression of LMPs,
BARTs, BARF1 and EBV-encoded microRNAs (latency types 0, I and II,
Table 2, Figure 3)[1, 15,17].
EBNA1 binds to oriP, the
latent origin of EBV replication that acts as a long-range enhancer of
Cp and LMP1p. This enhancer activity is essential for EBNA2 expression
during immortalization of B cells in
vitro [20].
In the absence of EBNA2 (the major viral transactivator protein in
latency type III), the LMP promoters are activated by cellular proteins
in nasopharyngeal carcinomas and in Sternberg-Reed cells of Hodgkin’s
disease (latency type II), but not in type I BL cell lines or BL
biopsies (type I latency) or memory B cells (type 0 latency,
characterized by variable Qp activity)[1].
It was observed by several laboratories that EBV latency
promoter-reporter constructs were active after in vitro transfection
into EBV carrying cell lines even if the corresponding latency promoter
located on the viral episomes was silent [21,22,23,24].
This observations
suggested that the transfected cells contained the transcription
factors necessary for promoter activity and raised the possibility that
silencing of latent viral promoters may be associated with their
inaccessibility to cellular transcription factors. In the next part we
briefly outline the epigenetic modifications and mechanisms potentially
regulating promoter accessibility.
Epigenetic modifications and
epigenetic regulatory systems:
Modified bases in the DNA of prokaryotes may serve as identification
marks that permit discrimination between self and foreign (invading)
DNA. Well characterized endo-deoxyribonuclease (restriction
endonuclease) and DNA methyltransferase (modification methylase)
activities, recognizing the same DNA sequence, enable bacterial cells
to resist infections by phage or plasmid DNA molecules that are
unmethylated or methylated at recognition sites different from those
marked on the host DNA (reviewed in Wilson, 1998, Bujnicki, 2001,
Murray, 2002). In addition to inhibiting the activity of restriction
endonucleases, sequence-specific methylation may modulate (repress or
activate) the expression of certain bacterial gene sets as well,
influencing thereby bacterial virulence [25,26].
Modified bases are also
involved in postreplicative repair processes in bacteria and may
contribute to membrane binding and segregation of the chromosome as
well [27,28].
The role of base modification mediated by DNA methyltransferases
apparently changed in large-genome eukaryotes. Bestor proposed that in
these organisms DNA methyltransferases perform a new function, the compartmentalization of the genome [29].
This would provide an easy access to the unmethylated fraction of the
genome by diffusible regulatory factors, while the usually larger,
methylated fraction would remain inaccessible for them. One could argue
that this novel function of DNA methyltransferases is reflected in the
discontinuous, and changing methylation patterns of vertebrate genomes.
In contrast to prokaryotes where the methyl-transferases modify
essentially all of their recognition sites, in vertebrates there are
alternating domains of methylated and unmethylated regions, and the
methylation patterns of a certain region may change during development
and differentiation (tissue-specific methylation patterns). The
unmethylated domains, called CpG islands, are regularly found 5’ to
coding sequences of constitutively expressed „housekeeping” genes [30,31].
It is important to note, however, that most CpG island promoters are
unmethylated independently of their activity [32,33].
Thus, mechanisms
other than DNA methylation may also silence CpG island promoters.
In mammals 5-methylcytosine (confined to the sequence 5’-CpG-3’,
shortly: CpG) is the predominant modified base found in DNA [34,35].
Comparison of the amino acid sequence of DNA methyltransferase I
(DNMT1), one of the mammalian DNA (cytosine-5)-methyltransferases, to
the sequences of bacterial methyltransferases suggested that DNMT1
arose by fusion of a prokaryotic modification methyltransferase gene
(coding for the C-terminal catalytic domain) and a second (unknown)
gene, coding for a regulatory (N-terminal) domain [36,37,38].
A key
feature of DNMT1 is its preference for hemimethylated DNA as a
substrate. Hemimethylated DNA molecules (consisting of one methylated
strand and an unmethylated complementary strand) are regularly produced
during DNA replication (provided that the parental strands were
originally methylated). High affinity of DNMT1 for hemimethylated DNA
ensures the clonal propagation of methylation patterns (maintenance
methylation). Other mammalian methyltransferases (like the human DNMT3A
and DNMT3B) create new methylation patterns by acting on unmethylated
DNA substrates (de novo
methylation)38. Their N-terminal domains differ from each other and
from that of DNMT1 [39].
Methylated CpG dinucleotides may silence promoters either directly, by
blocking the binding of transcription factors to their recognition
sequences, or indirectly, by attracting methyl-CpG binding proteins.
Methyl-CpG binding proteins regularly associate with histone
deacetylases and histone methylases. Removal of the acetyl moieties
from histone tails and methylation of selected lysine and arginine
groups results in a repressive chromatin structure silencing promoter
activity [40,41,42]. These alterations may switch
off active promoters and
stabilize the silent status of already inactivated genes. Riggs
proposed that stable gene silencing by DNA methylation may help progeny
cells to „remember” their proper cellular identity, i.e. DNA
methylation may provide a memory function [43].
Transcriptional repression may depend on CpG methylation density [33,44]. CpG-poor promoters
may retain their activity even if methylated. In somatic cells, a
second class of promoters characterized with a moderate or
intermediate CpG density (so called „weak
CpG island promoters”) are preferentially targeted by de novo methylation [33].
Typical CpG island promoters (called „strong
CpG island promoters”
by Weber et al.[33]) seem to be unmethylated
independent of their
activity [33] although they are regularly
inactivated by DNA methylation
in neoplasms [44]. Thus, focal hypermethylation
seems to accompany
tumorigenesis in parallel or consecutive to overall genomic
hypomethylation [45,46].
It is interesting to note, that an alternative system of epigenetic
memory, the Polycomb-trithorax group of protein complexes, that may
operate both independently from and in concert with DNA methylation,
ensures a heritable regulation of gene expression via modification of
histone tails, too (Bird, 2002, Cernilogar and Orlando, 2005, Laue et
al., 2005). In animal cells certain sets of promoters are marked by
trimethylation on lysine 27 of histone H3 (H3K27me3), a modification
carried out by the enzyme EZH2. EZH2, a component of the Polycomb
repressive complex 2 (PRC2) involved in gene silencing in embryonal
stem cells was recently shown to attract DNA methyltransferases47 and
may contribute, thereby, to permanent gene silencing via DNA
hypermethylation in cancer cells [48,49].
In contrast to the hypermethylated silent promoters, active promoters
are usually unmethylated at CpG dinucleotides and associate with
acetylated histones H3 and H4, that form ’acetylation islands’ [50].
Histone H3 di- or trimethylated on lysine 4 (H3K4me2, H3K4me3) is also
enriched at active promoters. The presence of these „activating”
chromatin modifications may not be sufficient, however, to ensure
promoter activity because certain promoters are enriched both in
„activating” and „repressive” modifications (’bivalent’ chromatin structure) [51]. Such ’bivalent’
domains may silence developmental genes in embryonic stem cells. In
addition, Weber et al. observed that unmethylated CpG rich promoters
may also associate with H3K4me2 in the absence of transcription [33].
Epigenetic mechanisms targeting
the alternative latency promoters Cp and Qp in lymphoma cells and
immortalized lympho-blastoid cell lines: The role of DNA
methylation in regulating Cp and Qp activity has been reviewed earlier
(Li and Minarovits, 2003). Briefly, both Cp and Qp can be silenced,
when inserted into reporter constructs, by in vitro CpG methylation.
Accordingly, Cp is hypermethylated and silent in tumor cells
corresponding to „Cp off”
latency (including latency type I Burkitt’s lymphoma cells and latency
type II Hodgkin’s lymphomas), but unmethylated and active in LCLs and
the majority of posttransplant lymphomas („Cp on” latency) [24,52, 53, 54]. Although in
vitro data
demonstrated thatCpG methylation could silence Qp as well [55],
Qp
stays unmethylated independently of its activity (i.e. it is
unmethylated also in LCLs where it is switched off). Thus, DNA
methylation may contribute to the silencing of Cp, but it is not used
to silence Qp in vivo.
Episomal EBV genomes are organized into chromatin [55].
Similarly to
cellular promoters [41, 56], the
activity of latent EBV promoters Cp and
Qp also correlated with certain histone modifications marking „open” or
„closed” chromatin domains [57,58,59,60]. In
lymphoblastoid cell lines
active C promoters were located to AcH3 and AcH4 rich regions60 that
are similar to the ’acetylation
islands’
characteristic to the active chromatin domains in human T cells [50]. In
contrast, in type I Burkitt’s lymphoma cell lines there was no
acetylation island at the inactive Cp. Active Qp, the promoter used in
most cases of „Cp off”
latency, was selectively enriched in AcH3, AcH4 and H3K4me2 [59,60] in
type I Burkitt’s lymphoma cell lines. In lymphoblastoid cell lines,
however, the inactive Qp was located to domains poor in acetylated
histones.
It was demonstrated by in vivo
footprinting that the unmethylated, active Cp was occupied by
transcription factors [53]. In contrast, the absence
of interactions with
activating transcription factors was characteristic for highly
methylated, silent C promoters. It is interesting to note, that
epigenetic silencing of Cp may be regarded as a viral escape mechanism,
because all of the transcripts encoding the immunodominant viral EBNA
proteins are initiated at Cp. Tumor cells actively using Cp can
survive, however, in severely immunsuppressed patients (e.g. B cell
lymphomas in transplantation recipients and AIDS patients).
We observed that the active, unmethylated Qp was also contacted by
cellular transcription factors and EBNA1, that in addition to oriP has
binding sites at Qp as well [53]. In contrast to Cp,
where silencing was
correlated with the loss of transcription factor binding, silent Qp was
marked by the same protein footprints as its active counterpart. A
unique footprint observed only at the silent Q promoters suggested that
binding of a cellular repressor might contribute to the repression of
Qp activity in „Cp on” latency.
Epigenetic marks at other EBV
latency promoters: In vitro
methylation experiments demonstrated that Wp, where transcripts of
EBNA1 to 6 are initiated soon after EBV infection of B cells (Figure
3), is a CpG methylation sensitive promoter61. Wp is switched off in
LCLs, however, provided that Cp is intact, in parallel with the
activation of Cp [62]. A variable methylation of Wp
in LCLs suggests that
epigenetic mechanisms other than CpG methylation also contribute to Wp
silencing [63].
The EBER 1 and 2 transcription units are transcribed by RNA polymerase
III. Their activity is also sensitive to in vitro
CpG methylation that blocks binding of the nuclear proteins c-myc and
ATF to the 5’region of EBER1p [64]. EBERs are
expressed in the major
EBV-carrying cell types and the EBER locus is hypomethylated [65].
LMP1 expression can be related to the methylation status of the LMP1
promoter. In type I BL lines LMP1p is silent and highly methylated,
whereas in LCLs and midline granulomas it is unmethylated and active
(reviewed by Li and Minarovits, 2003). In contrast to Cp, LMP1p
activity did not correlate with transcription factor occupancy [53].
In lymphoid cell lines, expression of LMP2A seems to be regulated by
the combinatorial effects of DNA methylation, histone acetylation and
the level of histone H3 dimethylated on lysine 4 (H3K4me2, a marker of
„open” chromatin)[66].
The promoter for the BARTs (also called CSTs including BARF0) RNAs was
found to be active and unmethylated in the only nasopharyngeal
carcinoma cell line it was looked for [67].
Latent Epstein-Barr virus
proteins interacting with the cellular epigenetic regulatory machinery:
We demonstrated above that various epigenetic control mechanisms leave
cell type specific marks on latent EBV genomes, establishing thereby
distinct viral epigenotypes (reviewed
by Minarovits,[68]). In turn, certain EBV encoded
proteins may interact
with the cellular epigenetic regulatory machinery, affecting thereby
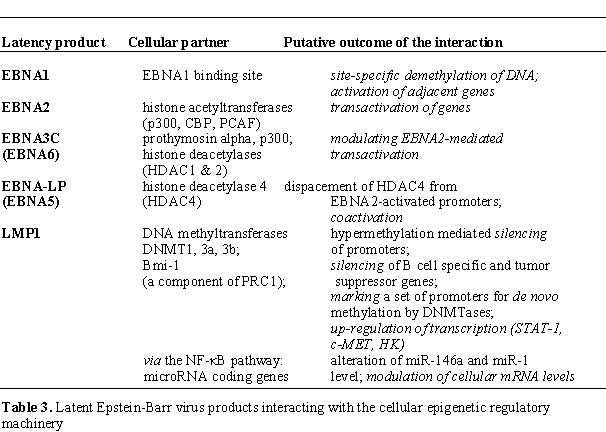
the activity of a set of cellular promoters (Table 3).
Because EBNA1 has binding sites in the
human genome [69] and it is known that high affinity
EBNA1 binding can
cause site specific demethylation within oriP, the latent origin of EBV
replication [70], we suggest that binding
of EBNA1 to its cellular target sites may elicit local demethylation
and contribute thereby to the activation of silent cellular promoters.
Lin et al. demonstrated in model experiments that high affinity of the
binding protein and protein binding site occupancy is crucial in
targeted demethylation [71].
EBNA2, the major viral transactivator protein expressed in „Cp on”
latency, may switch on both viral (Cp, LMP1p, LMP2Ap) and cellular
promoters (CD21p, CD23p, AML-2p, BATFp, IL-16p) (reviewed by Györy and
Minarovits [72]). EBNA2 activates its target
promoters through binding to
CBF1 (C promoter binding factor 1), a cellular protein that also
interacts with the activated cellular Notch family of proteins [73-76]. In vivo
footprinting confirmed CBF1 binding to the active C promoter and LMP2A
promoter but not to their inactive counterparts [53,77]. In addition to
CBF1, EBNA2 also associates with the cellular histone
acetyltransferases p300, CBP, and PCAF78. Wang et al.[78]
speculate that
these histone acetyltransferases may enhance the ability of EBNA2 to
up-regulate expression of the viral oncogene LMP1 by counteracting the
effect of histone deacetylases associated with the silent LMP1
promoter [78].
EBNA3C (EBNA6), a nuclear antigen essential for EBV-dependent
immortalization of B cells may fine tune (down-modulate) the
activity of EBNA2-activated promoters by interacting and competing with
prothymosin alpha, a partner of p300 [79,80].
In addition, EBNA3C
may also form complexes with histone deacetylases (HDAC1 and 2) in
human B cells [81]. Knight et al [81]
proposed that EBNA3C may be in separate
complexes with histone acetylases and deacetylases affecting the
function of distinct viral and cellular promoters.
In contrast to EBNA3C, the partly nuclear, parly cytoplasmic antigen
EBNA-LP (EBNA leader protein, EBNA5) strongly up-regulates
EBNA2-mediated transcription by relocalizing the histone deacetylase
HDAC4 from the nucleus to the cytoplasm [82].
Based on their
observations Portal et al. [82] suggested that the
coactivator function of
EBNA-LP is due to the reduction of the HDAC4 concentration in the
nucleus. They also speculated that EBNA-LP coactivates with EBNA2 by
displacing HDAC4 (and HDAC4 associated repressors) from EBNA2-bound
promoters. Recent data do not support, however, the nucleo-cytoplasmic
model for EBNA-LP mediated LMP1p coactivation, because an EBNA-LP
isoform, defective in shuttling to the cytoplasm, could coactivate
EBNA2 and associate with HDAC4 [83].
LMP1, a transmembrane protein involved in malignant transformation can
affect both alternative systems of epigenetic memory, DNA methylation
and the Polycomb-trithorax group of protein complexes. Tsai et al.
observed, however, that expression of the EBV-encoded oncoprotein LMP1
(latent membrane protein 1) in epithelial cells up-regulated the
expression and activity of cellular DNA methyltransferases 1, 3a and 3b
in vitro [84].
This
resulted in hypermethylation of the E-cadherin promoter and
down-regulation of E-cadherin gene expression. LMP1 activated DNMT1 via
the c-jun NH(2)-terminal kinase/activator protein-1 (JNK-AP-1)
signaling pathway [85]. In addition, formation of a
transcriptional
repression complex (composed of DNMT1 and histone deacetylase) could be
detected on the E-cadherin promoter as a consequence of LMP1 action.
How this repressor complex was targeted to the E-cadherin promoter,
however, remains to be elucidated. These results observed in epithelial
cells are certainly very much relevant to the development of
EBV-associated lymphomas as well, because latent EBV infection
apparently results in hypermethylation of a set of cellular promoters
not only in nasopharyngeal carcinoma [86] and
EBV-associated gastric
carcinoma [87,88], but also in typically LMP1
positive Hodgkin
lymphomas [89,90,91], iatrogenic lymphomas [92], and lymphomas developing in
AIDS patients [93].
Although the oncoprotein LMP1 can up-regulate the PcG protein Bmi-1 [94],
there are no data regarding the contribution of PcG (or trithorax
group) proteins to the regulation of latent EBV promoters.
Up-regulation of Bmi-1 is mediated by NF-κB signaling both in
EBV-positive and EBV-negative Hodgkin’s lymphoma cells. Dutton et al.94
suggested that by activating NF-κB, and thereby up-regulating Bmi-1,
LMP1 may contribute to the loss of B-cell identity in EBV carrying
Hodkins’s lymphomas via down-regulating a series of B-cell markers
(CD21/MS4A1, BLK, LY9). In EBV-negative disease other activators of
NF-κB may substitute for LMP1. We speculate that Bmi-1, as a component
of polycomb repressor complex 1 (PRC1), may act by marking a set of
promoters for de novo methylation by DNA methyltransferases. In
addition, Bmi-1, using the same mechanism, may down-regulate a series
of tumor suppressor genes (including IGSF4 and ATM), too. Bmi-1 also
mediates up-regulation of certain genes that are transcriptional
targets of LMP1 (STAT1 and c-MET and HK).
These genes code for signaling molecules and a hexokinase maintaining
high glycolytic activity and seem to play an important role in
lymphomagenesis.
It is worthy to mention that LMP1 may affect the levels of certain
cellular microRNAs (and modulate thereby cellular gene expression
programs) by activating the NF-κB pathway. LMP1 induced
expression of miR-146a, a microRNA that may affect the interferon
response (Cameron et al., 2008), and elevated the level of miR-155, a
microRNA implicated in the development of B cell lymphomas [95].
Stem cell chromatin patterns in
B-cell lymphomas: Burkitt lymphomas (BLs) and diffuse large
B-cell lymphomas (DLBCLs) seem to share a set of de novo methylated genes (e.g. HOXB13, CALCA, NEFL, PROK2)
that are repressed by the polycomb repressive complex 2 (PRC2) in
embryonic stem cells, in spite of the differences of BLs and DLBCLs
regarding morphology, genetic background, and transcriptional
pattern [96]. The aberrant DNA methylation observed
in lymphomas was
absent in adult stem and progenitor cells. Based on these data
Martin-Subero et al. [96] suggested that BLs and
DLBCLs may originate from
cells with stem cell features or acquire such features during
lymphomagenesis by epigenetic remodeling.
Tumor specific and
EBV-associated gene expression and CpG methylation changes in lymphomas:
The epigenetic alterations of the host genomes in EBV-associated
neoplasms (lymphomas and carcinomas) were reviewed recently [97].
It is obvious that certain common tumor suppressor and tumor-associated
genes were found to be silenced by DNA methylation in more than one
lymphoma types in independent studies. One could also discern, however,
methylation profiles that seem to be unique for individual lymphoma
types. The apparently specific contribution of EBV to the alteration of
gene expression in HD biopsies [98] remains to be
correlated with the
corresponding epigenetic changes. Elucidation of the temporal
sequence of genetic events (e.g. the Ig/c-myc translocation in
BL), epigenetic alterations (e.g. stem cell-like epigenetic marks of BL
cells) and EBV infection (an early event during the genesis of BL) will
be indispensable in case of each EBV positive lymphoma type for a
better understanding of lymphoma development and progression. In
addition, the patho-epigenetic changes induced by EBV, similarly to
those elicited by a growing number of other viruses and certain
bacteria may offer new opportunitis for therapeutic intervention.
References
- Niller, H.H., Wolf, H., Minarovits, J. (2007). Epstein-Barr Virus. In: Latency Strategies of Herpesviruses. Eds: Minarovits, J., Gonczol, E., Valyi-Nagy, T. Springer, pp.154-191.
- Shannon-Lowe, C.D., Neuhierl, B., Baldwin,
G., Rickinson, A.B., Delecluse, H. (2006). Resting B cells as a
transfer vehicle for Epstein-Barr virus infection of epithelial cells.
Proc. Natl. Acad. Sci. USA 103, 7065-7070.

- Epstein, M.A., Achong, B.G., Barr, Y.M. .
Virus particles in cultured lymphoblasts from Burkitt’s
lymphoma. Lancet 1964; 1, 702-703.

- Li, H., Minarovits, J. Host
cell-dependent expression of latent Epstein-Barr virus genomes:
Regulation by DNA methylation. Adv
Cancer Res. 2003;89:133-56..

- Dillner, J., Kallin, B. The
Epstein-Barr virus proteins. Adv
Cancer Res. 1988;50:95-158.

- Hammerschmidt, W., Sugden., B.
Identification and characterization of orilyt, a lytic origin of DNA
replication of Epstein-Barr virus. Cell.
1988 Nov 4;55:427-33.

- Sato, H., Takimoto, T., Tanaka, S., Tanaka,
J., Raab-Traub, N. Concatemeric replication of Epstein-Barr
virus: structure of termini in virus producer and newly transformed
cell lines, J. Virol.1990;64,5295-5300.

- Fixman, E.D., Hayward, G.S., Hayward D.
Trans-acting requirements for replication of Epstein-Barr virus
ori-Lyt. Virology 1992;66, 5030-5029.

- Yates, J.L., Warren, N., Reisman, D.,
Sugden, B. A cis-acting element from the Epstein-Barr virus
genome that permits stable replication of recombinant plasmids in
latently infected cells. Proc. Natl. Acad. Sci. USA 1984; 81, 3806-3810.

- Nanbo, A., Sugden, A., Sugden, B.
The coupling of synthesis and partitioning of EBV’s plasmid replicon is
revealed in live cells. EMBO J.2007; 26, 4252-4262.

- Raab-Traub, N., Flynn, K. The
structure of the termini of the Epstein-Barr virus is a marker of
clonal cell proliferation, Cell 1986;47, 883-889.

- Miller, G. (1990). Epstein-Barr virus: Biology, pathogenesis and medical aspects. In: Virology. Eds: B.N. Fields, D.M. Knipe. Raven Press, New York, pp. 1921-1958.
- Niller, H.H., Salamon, D., Banati, F.,
Schwarzmann, F., Wolf, H., Minarovits, J. (2004). The LCR of EBV
makes Burkitt’s lymphoma endemic, Trends Microbiol 12, 495-499.

- Contreras-Brodin BA, Anvret M, Imreh S,
Altiok E, Klein G, Masucci MG. B cell phenotype-dependent expression of
the Epstein-Barr virus nuclear antigens EBNA-2 to EBNA-6: studies
with somatic cell hybrids. J Gen Virol. 1991;72 :3025-33.

- Smith, P. Epstein-Barr virus complementary
strand transcripts (CSTs/BARTs) and cancer. Semin. Cancer Biol.
2001;11, 469-476.

- Pfeffer S., Zavolan M., Grässer F.A.,
Chien M., Russo J.J., Ju J., John B., Enright A.J., Marks D.,
Sander C. Identification of virus-encoded microRNAs. Science
2004;314, 734-736.

- Cai, X., Schafer, A., Lu, S.,
Bilello, J.P., Desrosiers, R.C., Edwards, R., Raab-Traub,
N., Cullen B.R. . Epstein-Barr virus microRNAs are evolutionarily
conserved and differentially expressed. PLoS Pathog. 2006; 2, e23.

- Grundhoff, A., Sullivan, C.S., Ganem, D. A
combined computational and microarray-based approach
identifies novel microRNAs encoded by human gamma-herpesviruses.
RNA 2006;12, 733-750.

- Swaminathan, S. Noncoding RNAs produced by
oncogenic human herpesviruses. J. Cell. Physiol. 2008 216, 321-326.

- Altmann, M., Pich, D., Ruiss, R., Wang,
J., Sugden, B. . Transcriptional activation by EBV nuclear
antigen 1 is essential for the expression of EBV’s transforming genes.
Proc Natl. Acad. Sci. USA 2006;103,14188-14193.

- Woisetschlaeger, M., Jack, L., Strominger,
J.L., Speck, S. Mutually exclusive usage of viral promoters in
Epstein-Barr virus latently infected lymphocytes. Proc. Natl. Acad.
Sci. USA 1989;86, 6498-6502.

- Walls, D., Perricaudet, M. Novel
downstream elements upregulate transcription initiated from an
Epstein-Barr virus latent promoter. EMBO J.1991; 10, 143-151.

- Minarovits, J., Hu, L.F.,
Minarovits-Kormuta, S., Klein, G., Ernberg, I. Sequence-specifc
methylation inhibits the activity of the Epstein-Barr virus LMP1 and
BCR2 enhancer-promoter regions. Virology 1994; 200, 661-667.

- Robertson, K.D., Hayward, D., Ling, P.D.,
Samid, D., Ambinder, R.Transcriptional activation of the
Epstein-Barr virus latency C promoter after 5-azacytidine treatment:
evidence that demethylation at a single CpG site is crucial. Mol. Cell.
Biol.1995; 15, 6150-6159 .

- Heithoff, D.M., Sinsheimer, R.L., Low,
D.A., Mahan, M.J. An essential role for DNA adenine methylation
in bacterial virulence. Science 1999; 284, 967-970.

- Balbontin, R., Rowley, G., Pucciarelli,
M.G., Lopez-Garrido, J., Wormstone, Y., Lucchini, S., Garcia-Del
Portillo, F., Hinton, J.C., Casadesus, J. DNA adenine
methylation regulates virulence gene expression in Salmonella enterica
serovar Typhimurium. J Bacteriol 2006;188, 8160-8168.

- Pukkila, P.J. Telling right from wrong: a role for DNA methylation. Trends in Genetics 1987; 3, 1-2.
- Ogden, G.B., Pratt, M.J., Schaechter,
M. The replicative origin of the E. coli chromosome binds to cell
membranes only when hemimethylated. Cell 1988;54, 127-135.

- Bestor, T.H. DNA methylation:
evolution of a bacterial immune function into a regulator of gene
expression and genome structure in higher eukaryotes. Phil Trans R Soc
Lond B 1990;326, 179-187.

- Bird, A.P. CpG islands and the function of
DNA methylation. Nature 1986; 321, 209-213

- Bird, A.P .CpG islands as gene markers in the vertebrate nucleus. Trends in Genetics 1987; 3, 342-347.
- Bird, A.DNA methylation patterns and
epigenetic memory. Genes Dev 2002;16, 6-21.

- Weber, M., Hellmann, I., Stadler, M.B.,
Ramos, L., Pääbo, S., Rebhan, M, Schübeler, D.. Distribution,
silencing potential and evolutionary impact of promoter DNA methylation
int he human genome. Nature Genetics 2007;39, 457-466.

- Razin, A., Riggs, A.D.DNA methylation and
gene function. Science 1980;210, 604-610.

- Paulsen, M., Tierling, S., Walter, J. DNA methylation in the mammalian genome. In: Epigenetics. Ed.: Trost, J. Caister Academic Press, 2008 Norfolk, UK, pp.1-21.
- Bestor, T., Laudano, A., Mattaliano, R.,
Vernon, I. Cloning and sequencing of a cDNA encoding DNA
methyltransferase of mouse cells. J Mol Biol 1988;203, 971-983.

- Yen, R.W.C., Vertino, P.M., Nelkin, B.D.,
Yu, I.J., Eldeiry, W., Cumarraswamy, A., Lennon, G.G., Trask, B.J.,
Celano, P., Baylin, S.B.Isolation and characterization of the
cDNA encoding human DNA methyltransferase. Nucleic Acids Res. 1992;20,
2287-2291.

- Van Emburgh, B.O., Robertson, K.D. DNA methyltransferases and methyl-CpG binding proteins as multifunctional regulators of chromatin structure and development. In: Epigenetics. Ed.: Trost, J. Caister Academic Press, Norfolk,2008 UK, pp. 23-61.
- Xie, S., Wang, Z., Okano, M., Nogami, M.,
Li, Y., He, W.W., Okumura, K., Li, E. Cloning, expression and
chromosome locations of the human DNMT3 gene family. Gene 1999;236,
87-95.

- Jones, P.L., Veenstra, G.J., Wade, P.A.,
Vermaak, D., Kass, S.U., Landsberger, N., Strouboulis, J.,
Wolffe, A.P. . Methylated DNA and MeCP2 recruit histone
deacetylase to repress transcription. Nat Genet 1998;19, 187-191.

- Jenuwein, T., Allis, C.D.
Translating the histone code. Science 2001;293, 2889-2897.

- Fuks, F., Hurd, P.J., Wolf, D., Nan, X.,
Bird, A.P., Kouzarides, T. The methyl-CpG-binding protein MeCP2
links DNA methylation to histone methylation. J Biol Chem 2003; 278,
4035-4040.

- Riggs, A.D. DNA methylation and
late replication probably aid cell memory, and type I DNA reeling could
aid chromosome folding and enhancer function. Phil Trans R Soc Lond B
1990; 326,285-297.

- Baylin SB, Herman JG. DNA hypermethylation
in tumorigenesis: epigenetics joins genetics. Trends Genet. 2000
16:168-74.

- Ehrlich, M. DNA hypomethylation and cancer. In: DNA Alterations in Cancer. Ed.: Ehrlich, M. Eaton publishing, Natick, MA. pp. 273-291.
- Zilberman, D.The human promoter methylome.
Nature Genetics 2007;39, 442-443.

- Vire, E., Brenner, C., Deplus, R.,
Blanchon, L., Fraga, M., Didelor, C., Morey, L., Van Eynde, A.,
Bernard, D., Vanderwinden, J.M., Bollen, M., Esteller, M., Di Croce,
L., de Launoit, Y., Fuchs, F. The Polycomb group protein EZH2
controls DNA methylation. Nature 2006; 439, 871-874.

- Schlesinger, Y., Straussman, R., Keshet,
I., Farkash, S., Hecht, M., Zimmerman, J., Eden, E., Yakhini, Z.,
Ben-Shushan, E., Reubinoff, B., Bergman, Y., Simon, I., Cedar, H.
Polycomb-mediated methylation on Lys27 of histone H3 pre-marks
genes for de novo methylation in cancer. Nature Genetics 2007; 39,
232-236.

- Ohm, J.E., McGarvey, K.M., Yu, X., Cheng,
L., Schuebel, K.E., Cope, L., Mohammad, H., Chen, W., Daniel, V.C., Yu,
W., Berman, D.M., Jenuwein, T., Pruitt, K., Sharkis, S.J., Watkins,
D.N., Herman, J.G., Baylin, S. (2007). A stem cell-like chromatin may
predispose tumor suppressor genes to DNA hypermethylation and heritable
silencing. Nature Genetics 39, 237-242.

- Roh, T.Y., Cuddapah, S., Zhao, K.Active
chromatin domains are defined by acetylation islands defined by
genome-wide mapping. Genes Dev. 2005;19, 542-555.

- Bernstein, B.E., Mikkelsen, T.S., Xie, X.,
Kamal, M., Huebert, D.J., Cuff, J., Fry, B., Meissner, A., Wernig,
Plath, K., Jaenisch, R., Wagschal, A., Feil, R., Schreiber, S.L.,
Lander, E.S.A bivalent chromatin structure marks key
developmental genes in embryonic stem cells. Cell 2006;125, 315-326.

- Robertson, K.D., Manns, A., Swinnen, L.J.,
Zong, J.C., Gulley, M.L., Ambinder, R.F.CpG methylation of the
major Epstein-Barr virus latency promoter in Burkitt’s lymphoma and
Hodgkin’s disease. Blood 1996;88, 3129-3136.

- Salamon D., Takacs M., Ujvari, D., Uhlig,
J., Wolf, H., Minarovits, J., Niller, H.H. Protein-DNA-binding
and CpG methylation at nucleotide resolution of latency-associated
promoters Qp, Cp and LMP1p of Epstein-Barr virus, J. Virol. 2001; 75,
2584-2596.

- Tao, Q., Robertson, K.D., Manns, A.,
Hildesheim, A., Ambinder, R.F. The Epstein-Barr virus major
latent promoter Qp is constitutively active, hypomethylated and
methylation sensitive. J. Virol.1998; 72, 7075-7083.

- Dyson, P., Farrell, P.J. (1985). Chromatin
structure of Epstein-Barr virus, J. Gen. Virol. 66, 1931-1940.

- Verdone, L., Caserta, M., Di Mauro, E.
Role of histone acetylation in the control of gene expression.
Biochem. Cell Biol.2005;83, 344-353.

- Alazard, N., Gruffat, H., Hiriart, E.,
Sergeant, A., Manet, E. Differential hyperacetylation of
histones H3 and H4 upon promoter-specific recruitment of EBNA2 in
Epstein-Barr virus chromatin. J. Virol. 2003; 77, 8166-8172.

- Chau, C.M., Lieberman, P.M.Dynamic
chromatin boundaries delineate a latency control region of
Epstein-Barr virus, J. Virol. 2004;78, 12308-12319.

- Day, L., Chau, C.M., Nebozhyn, M.,
Rennekamp, A.J., Showe, M., Lieberman, P.M. Chromatin profiling
of Epstein-Barr virus latency control region, J. Virol. 2007; 81,
6389-6401.

- Fejer, G., Koroknai, A., Banati, F.,
Györy, I., Salamon, D., Wolf H., Niller H.H., Minarovits, J.
Latency type-specific distribution of epigenetic marks at the
alternative promoters Cp and Qp of Epstein-Barr virus, J. Gen. Virol.
2008;
89, 1364-1370.

- Jansson, A., Masucci, M., Rymo, L.
Methylation at discrete sites within the enhancer region regulatest he
activity of the Epstein-Barr virus BamHI W promoter in Burkitt lymphoma
cell lines. J. Virol.1992; 66, 62-69.

- Woisetschlaeger M, Jin XW, Yandava CN,
Furmanski LA, Strominger JL, Speck SH. Role for the Epstein-Barr virus
nuclear antigen 2 in viral promoter switching during initial stages of
infection. Proc Natl Acad Sci U S A. 1991;88:3942-6.

- Elliott, J., Goodhew, E.B., Krug, L.T.,
Shaknowsky, N., Yoo, L., Speck, S.H. (2004). Variable methylation of
the Epstein-Barr virus Wp EBNA gene promoter in B-lymphoblastoid cell
lines. J. Virol. 78, 14062-14065.

- Banati, F., Koroknai, A., Salamon, D.,
Takacs, M., Minarovits-Kormuta, S., Wolf, H., Nimmer, H.H., Minarovits,
J. (2008). CpG-methylation silences the activity of the RNA polymerase
III transcribed EBER-1 promoter of Epstein-Barr virus. FEBS Letters
582, 705-709.

- Minarovits, J., Hu, L.F., Marcsek, Z.,
Minarovits-Kormuta, S., Klein, G., Ernberg, I. RNA polymerase
III-transcribed EBER 1 and 2 transcription units are expressed and
hypomethylated in the major Epstein-Barr virus-carrying cell types. J.
Gen. Virol. 1992 73, 1687-1692.

- Gerle, B., Koroknai, A., Fejer, G., Bakos,
A., Banati, F., Szenthe, K., Wolf, H., Niller, H.H., Minarovits, J.,
Salamon, D.Acetylated histone H3 and H4 mark the upregulated
LMP2A promoter of Epstein-Barr virus in lymphoid cells. J. Virol.
2007;81,
13242-13247.

- de Jesus, O., Smith, P.R., Spender, L.C.,
Elgueta, K.C., Niller, H.H., Huang, D., Farrell, P.J. Updated
Epstein-Barr virus (EBV) DNA sequence and analysis of a promoter for
the BART (CST, BARF0) RNAs of EBV. J. Gen. Virol. 2003;84, 1443-1450.

- Minarovits, J. Epigenotypes of latent
herpesvirus genomes. Current Topics Microbiol. Immunol.2006; 310, 61-80.

- Dresang, L.R., Vereide, D.T., Sugden,
B.Identifying sites bound by Epstein-Barr virus nuclear antigen 1
(EBNA1) int he human genome: defining a position-weighted matrix to
predict sites bound by EBNA1 in viral genomes. J. Virol. 2009;83,
2930-2940.

- Hsieh, C.L. Evidence that protein binding
specifies sites of DNA demethylation. Mol. Cell. Biol. 1999; 19, 46-56.

- Lin, I.G., Tomzynski, T.J., Ou, Q, Hsieh,
C.L. Modulation of DNA binding protein affinity directly
affects target site demethylation. Mol. Cell. Biol. 2000; 20, 2343-2349

- Györy I, Minarovits J. Epigenetic
regulation of lymphoid specific gene sets. Biochem Cell Biol. 2005
Jun;83(3):286-95

- Ling, P.D., Hsieh, J.D., Ruf,
I.K., Rawlins, D.E., Hayward S.D. EBNA-2 upregulation of
Epstein-Barr virus promoters and the cellular CD23 promoter utilizes a
common targeting intermediate. CBF1. J. Virol. 1994 68, 5375-5383.

- Hsieh, J.J.D., Hayward, S.D.
Masking the CBF1/RBPJκ transcriptional repression domain by
Epstein-Barr virus EBNA2. Science 1995;268, 560-563.

- Hsieh, J.J.D., Nofziger, D., Weinmaster,
G., Hayward, D. Epstein-Barr virus immortalization: Notch2
interacts with CBF1 and blocks differentiation. J. Virol. 1997; 71,
1938-1945.

- Höfelmayr, H., Strobl, L.J., Marschall,
G., Bornkamm, G.W., Zimmer-Strobl, U. (2001). Activated Notch 1 can
transiently substitute for EBNA2 in the maintenance of proliferation of
LMP1-expressing immortalized B cells. J. Virol. 75, 2033-2040.

- Salamon, D., Takacs, M., Schwarzmann, F.,
Wolf, H., Minarovits, J., Niller, H.H. (2003). High-resolution
methylation analysis and in vivo protein-DNA binding at the promoter of
the viral oncogene LMP2A in B cell lines carrying latent Epstein-Barr
virus genomes. Virus Genes 27, 57-66.

- Wang, L., Grossman, S.R., Kieff, E.
Epstein-Barr virus nuclear protein 2 interacts with p300, CBP,
and PCAF histone acetyltransferases in activation of the LMP1 promoter.
Proc. Natl. Acad. Sci. USA 2000; 97, 430-435.

- Cotter II, M.A., Robertson, E.S.Modulation
of histone acetyltransferase activity through interaction of
Epstein-Barr Nuclear Antigen 3C with prothymosin alpha. Mol. Cell.
Biol. 2000;20, 5722-5735.

- Subramanian, C., Hasan, S., Rowe, M.,
Hottinger, M., Orre, R., Robertson, E.S. Epstein-Barr virus
nuclear antigen 3C and prothymosin alpha interact with the p300
transcriptional coactivator at the CH1 and CH3/HAT domains and
cooperate in regulation of transcription and histone acetylation. J.
Virol.2002;76, 4699-4708.

- Knight JS, Lan K, Subramanian C, Robertson
ES. Epstein-Barr virus nuclear antigen 3C recruits histone deacetylase
activity and associates with the corepressors mSin3A and NCoR in human
B-cell lines. J Virol. 2003 ;77 4261-72.

- Portal D., Rosendorff, A., Kieff, E.
(2006). Epstein-Barr nuclear antigen leader protein coactivates
transcriotion through interaction with histone deacetylase 4. Proc.
Natl. Acad Sci USA 103, 19278-19283.

- Ling PD, Tan J, Peng R.
Nuclear-cytoplasmic shuttling is not required for the
Epstein-Barr virus EBNA-LP transcriptional coactivation function. J
Virol. 2009 ;83:7109- 29.

- Tsai, C.N., Tsai, C.L., The, K.P., Chang,
H.Y., Chang, Y.S. The Epstein-Barr virus oncogene product,
latent membrane protein 1, induces the down-regulation of E-cadherin
gene expression via activation of DNA methyltransferases. Proc. Natl.
Acad. Sci. USA 2002; 99, 10084-10089.

- Tsai, C.L., Li, H.P., Lu, Y.L., Hsueh, C., Liang, Y., Chen, C.L., Tsao, S.W., The, K.P., Yu, J.S., Chang, Y.S. Activation of DNA methyltransferase 1 by EBV LMP1 involves c-Jun NH2-terminal kinase signaling. Cancer Res.2006; 66, 11668-11676.
- Kwong, J., Lo, K.W., To, K.F., Teo, P.M.,
Johnson, P.J., Huang, D.P. Promoter hypermethylation of
multiple genes in nasopharyngeal carcinoma. Clin Cancer Res 2002;8,
131-137.

- Kang, G.H., Lee, S., Kim, W.H., Lee, H.W.,
Kim, J.C., Rhyu, M.G., Ro, J.Y. Epstein-Barr virus-positive
gastric carcinoma demonstrates frequent aberrant methylation of
multiple genes and constitutes CpG island methylator phenotype-positive
gastric carcinoma. Am. J. Pathol. 2002;160, 787-794.

- Chang, M.S., Uozaki, H., Chong, J.M.,
Ushiku, T., Sakuma, K., Ishikawa, S., Hino, R., Barua, R.R., Iwasaki,
Y., Arai, K., Fujii, H., Nagai, H., Fukayama, M. CpG island
methylation status in gastric carcinoma with and without infection of
Epstein-Barr Virus. Clin. Cancer Res. 2006; 12, 2995-3002.

- Murray, P.G., Qiu, G.H., Fu, L., Waites,
E.R., Srivastava, G., Heys, D., Agathanggelou, A., Latif, F., Grundy,
R.G., Mann, J.R., Straczynsky, J., Crocker, J., Parkes, S.E., Ambinder,
R.F., Young, L.S., Tao, Q. (2004). Frequent inactivation of the RASSF1A
tumor suppressor gene in Hodgkin’s lymphoma. Oncogene 23, 1326-1331.

- Doerr JR, Malone CS, Fike FM, Gordon MS,
Soghomonian SV, Thomas RK, Tao Q,Murray PG, Diehl V, Teitell MA, Wall
R. Patterned CpG methylation of silenced B cell gene promoters in
classical Hodgkin lymphoma-derived and primary effusion lymphoma cell
lines. J Mol Biol. 2005 ;350:631-40

- Ushmorov A, Leithäuser F, Sakk O,
Weinhaüsel A, Popov SW, Möller P, Wirth T. Epigenetic processes play a
major role in B-cell-specific gene silencing in classical Hodgkin
lymphoma. Blood. 2006 ;107:2493-500.

- Au, W.Y., Ma, E.S., Choy, C., Chung, L.P.,
Fung, T.K., Liang, R., Kwong, Y.L. . Therapy-related lymphomas in
patients with autoimmune diseases after treatment with
disease-mofifying anti-rheumatic drugs. Am. J. Hematol.2006; 81. 5-11.

- Rossi D., Gaidano G., Gloghini, A.,
Deambrogi, C., Franceschetti S., Berra, E., Cerri, M., Vendramin, C.,
Conconi, A., Viglio, A., Muti, G., Oreste, P., Morra, E., Paulli, M.,
Capello, D., Carbone, A. Frequent aberrant promoter
hypermethylation of O6-methylguanine-DNA methyltransferase and
death-associated protein kinase genes in immunodeficiency-related
lymphomas. Br. J. Haematol. 2003; 123, 475-478.

- Dutton, A., Woodman, C.B., Chukwuma,
M.B., Wei, W., Vockerodt, M., Baumforth, K.R., Flavell, J.R.,
Taylor, A.M., Young, L.S., Murray, P.G. Bmi-1 is induced by the
Epstein-Barr virus viral oncogene LMP-1 and regulates the expression of
target genes in Hodgkin lymphoma cells. Blood 2007; 109, 2597-2603.

- Lu, F., Weidmer, A., Liu, C.G., Volinia,
S., Croce, C.M., Lieberman, P.M.Epstein-Barr virus-induced
miR-155 attenuates NF-κB signaling and stabilizes latent virus
persistence. J. Virol. 2008;82, 10436-10443.

- Martin-Subero, J., Kreuz, M., Bibikova,
M., Bentink, S., Ammerpohl, O., Wickham-Garcia, E., Rosolowski, M.,
Richter, J., Lopez-Serra, L., Ballestar, E., Berger, H., Agirre, X.,
Bernd, H.W., Calvanese, V., Cogliatti, S.B., Drexler, H.G., Fan, J.B.,
Fraga, M.F., Hansmann, M.L., Hummel, M., Klapper, W., Korn, B.,
Küppers, R., MacLeod, R.A.F., Möller, P., Ott, G., Pott, C., Prosper,
F., Rosenwald, A., Schwaenen, C., Schübeler, D., Seifert, M.,
Stürzenhofecker, B., Weber, M., Wessendorf, S., Loeffler, M., Trümper,
L., Stein, H., Spang, R., Esteller, M., Barker, D., Hasenclever, D.,
Siebert, R. New insights into the biology and origin of mature
agressive B-cell lymphomas by combined epigenomic, genomic, and
transcriptional profiling. Blood 2009; 113, 2488-2497.

- Niller, H.H., Wolf, H., Minarovits.
J.Epigenetic dysregulation of the host cell genome in
Epstein-Barr virus-associated neoplasia. Seminars in Cancer Biology 19,
158-164. Noyer-Weidner, M., Jentsch, S.,Pawlek, B., Günthert, U.,
Trautner, T.A. . Restriction and modification in Bacillus
subtilis: DNA methylation potential of the related bacteriophages Z,
SPR, SP beta phi 37 and rho 11. J Virol 2009;1983;46, 446-453.

- Garcia, J.F., Camacho, F.I., Morente, M.,
Fraga, M., Montalban, C., Alvaro, T., Bellas, C., Castano, A., Diez,
A., Flores, T., Martin, C., Martinez, M.A., Mazorra, F., Menárguez, J.,
Mestre, M.J., Mollejo, M., Saez, A.I., Sanchez, L., Piris, M.A.,
Spanish Hodgkin Lymphoma Study Group. Hodgkin and
Sternberg-Reed cells harbor alterations int he major tumor suppressor
pathways and cell-cycle checkpoints: analysis using tissue microarrays.
Blood 2003; 101, 681-689.