Genetic Pathways Leading to Therapy-Related Myeloid Neoplasms
University of
Chicago, Chicago, IL, USA
Correspondence
to: Michelle M. Le Beau, PhD, University of Chicago, 5841 S.
Maryland Avenue,
MC2115, Chicago, Il 60637. Telephone: +1
773-702-0795, Fax: +1 773-702-9311.
E-mail: mlebeau@bsd.uchicago.edu
Published: May 16, 2011
Received: March 18, 2011
Accepted: April 21, 2011
Mediterr J Hematol Infect Dis 2011, 3: e2011019, DOI 10.4084/MJHID.2011.019
This article is available from: http://www.mjhid.org/article/view/8226
This is an Open Access article
distributed under the terms of the
Creative Commons Attribution License
(http://creativecommons.org/licenses/by/2.0),
which permits
unrestricted use, distribution, and reproduction in any medium,
provided the original work is properly cited
Abstract
Therapy-related
myeloid neoplasm (t-MN) is a distinctive clinical syndrome occurring
after exposure to chemotherapy or radiotherapy. t-MN arises in
most cases from a multipotential hematopoietic stem cell or, less
commonly, in a lineage committed progenitor cell. The prognosis
for patients with t-MN is poor, as current forms of therapy are largely
ineffective. Cytogenetic analysis, molecular analysis and gene
expression profiling analysis of t-MN has revealed that there are
distinct subtypes of the disease; however, our understanding of the
genetic basis of t-MN is incomplete. Elucidating the genetic
pathways and molecular networks that are perturbed in t-MNs, may
facilitate the identification of therapeutic targets that can be
exploited for the development of urgently-needed targeted therapies.
Introduction
t-MN is a late complication of cytotoxic therapy (radiation and/or chemotherapy) used in the treatment of both malignant and non-malignant diseases.[1-4] Several distinct cytogenetic and clinical subtypes of t-MN are recognized that are closely associated with the nature of the preceding treatment. Patients who develop t-MN following alkylating agent therapy (~75% of cases) typically show a latency of 3-7 years from alkylating agent exposure (median 5 years), insidious disease onset with an antecedent MDS with peripheral cytopenias, loss, deletion, or rearrangement of chromosomes 5 and/or 7, and a poor prognosis (median survival <8 months).[1] Typically, all three hematopoietic cell lineages (erythroid, myeloid, and megakaryocytic) are involved in the dysplastic process (trilineage dysplasia), suggesting that the disease arises in a multipotent hematopoietic stem or progenitor cell (HSPC). In contrast, patients who develop t-MN following treatment with drugs targeting topoisomerase II are younger, have a shorter latency period (2-3 years), and present with AML. Balanced translocations involving MLL at 11q23, RUNX1 at 21q22, CBFB at 16q22, or PML (15q24) and RARA (17q12) are common in this subgroup, suggesting that these cytogenetic subsets of t-MN arise in a lineage committed progenitor cell.[3-5] Survival times of t-MN patients are often short, because this disorder is less responsive to current forms of therapy than is AML de novo.[1,3,4]
t-MN represents an important model for cancer. The incidence of t-MN is rising, as a result of the increasing number of cancer survivors at risk of developing this disorder and the changes in therapeutic trends. Secondly, t-MN provides a unique opportunity to examine the effects of mutagens on carcinogenesis in humans, as well as the role of genetic susceptibility to cancer.[3] Finally, the mechanisms of leukemogenesis that are uncovered in t-MN will likely apply to those subtypes of AML de novo, which share the same cytogenetic abnormalities, e.g., AML de novo with abnormalities of chromosome 5 or 7. In this chapter, we review the genetic characteristics of t-MN with an emphasis on defining the genetic pathways leading to t-MN with a del(5q).
Cytogenetic Analyses
Table 1 summarizes the cytogenetic pattern in the recently updated University of Chicago series of 386 consecutive patients with t-MN. Of these, 349 (90.4%) had a clonal chromosomal abnormality, including 259 (67%) with a clonal abnormality leading to loss, deletion, or rearrangements of chromosomes 5 and/or 7 (referred to as del(5q)/t(5q) and -7/del(7q) herein),4 (Le Beau and Larson, unpublished data). Overall, 164 patients (42%) had abnormalities of chromosome 5, and 180 (47%) had abnormalities of chromosome 7. Of these patients, eighty-five patients had abnormalities of both chromosomes 5 and 7. A del(5q) was the most common structural abnormality. The pattern of numerical and structural abnormalities is shown in Figure 1, and illustrates that t-MN is associated with a complex karyotype, with a predominance of the loss of genetic material.
In other studies, we evaluated the cytogenetic pattern of 3,444 patients with primary MDS, AML de novo, or t-MN evaluated over the past 35 years by our Cancer Cytogenetics Laboratory. Of these, 553 (16%) patients had del(5q)/t(5q), and 597 (17.3%) had -7/del(7q) (Le Beau et al., unpublished data). Complex karyotypes were associated with abnormalities of chromosome 5, rather than chromosome 7. Recurring abnormalities observed at a high frequency (>20%) in patients with del(5q) included +8, and loss of 13q, 16q, 17p (40% of cases), chromosome 18, and 20q, which frequently occured in the same clone (Figure 1, and data not shown).
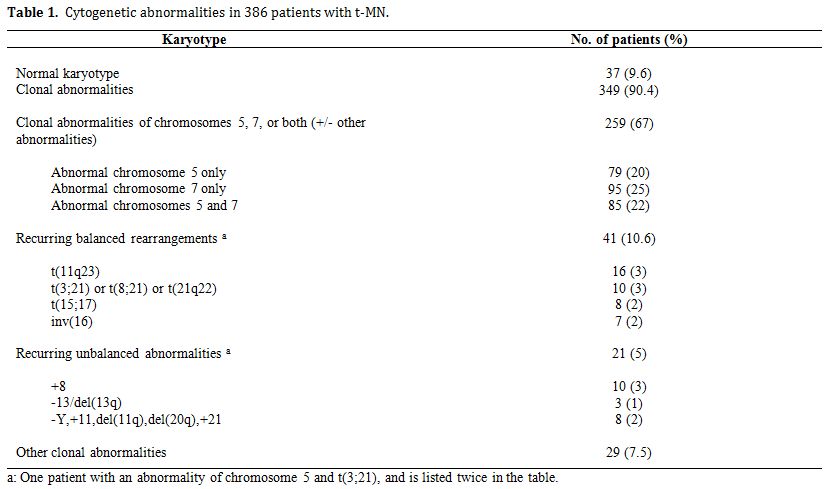
Table 1. Cytogenetic abnormalities in 386 patients with t-MN.
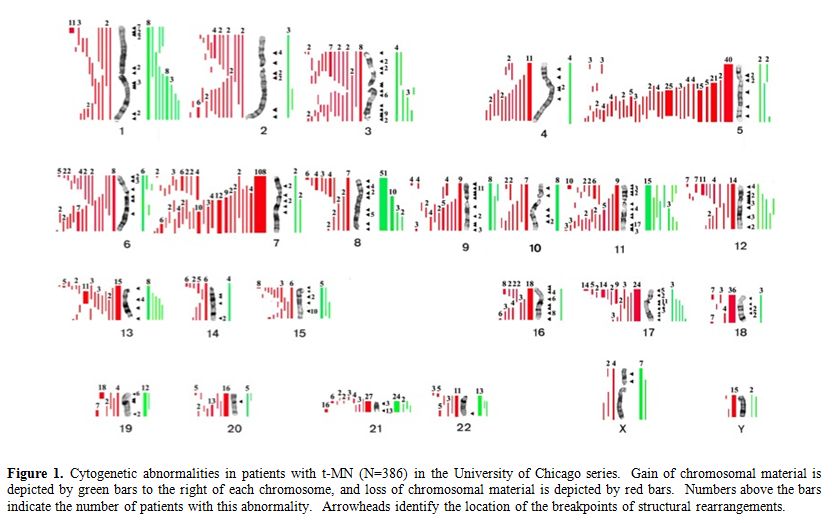
Figure 1. Cytogenetic abnormalities in patients with t-MN (N=386) in the University of Chicago series. Gain of chromosomal material is depicted by green bars to the right of each chromosome, and loss of chromosomal material is depicted by red bars. Numbers above the bars indicate the number of patients with this abnormality. Arrowheads identify the location of the breakpoints of structural rearrangements.
Identification of Haploinsufficient Myeloid Suppressor Genes on 5q
Several groups of investigators have defined a commonly deleted segment (CDS) on the long arm of chromosome 5 predicted to contain a myeloid tumor suppressor gene. [6-8] Using cytogenetic and molecular analysis of MDS, AML and t-MN with a del(5q), we previously defined a region of 970 kilobase within 5q31.2, flanked by D5S479 and D5S500, that is deleted in all patients (Figure 2), and determined the genomic sequence of this region.6,9 In subsequent studies, we generated a transcript map of the CDS, and identified and cloned 19 genes;9 the CDS also contains one miRNA. The functions of the proteins encoded by these genes are diverse, and include the regulation of mitosis and the G2 checkpoint, transcriptional control, and translational regulation.
MDS with an isolated del(5q) (the 5q- Syndrome) is a distinct subtype of MDS, characterized by a macrocytic anemia, female predominance, and a favorable outcome, with a low risk of transformation to AML.10 Boultwood and colleagues identified a 1.5 Mb CDS within 5q33.1 between D5S413 and GLRA1 .8 This region is distal to the CDS in 5q31.2 found in patients with AML with a del(5q). In summary, the existing data suggest that there are two non-overlapping CDSs: 5q31.2 in the more aggressive form of MDS, AML de novo, and t-MN, and 5q33.1 in the 5q- Syndrome (Figure 2).
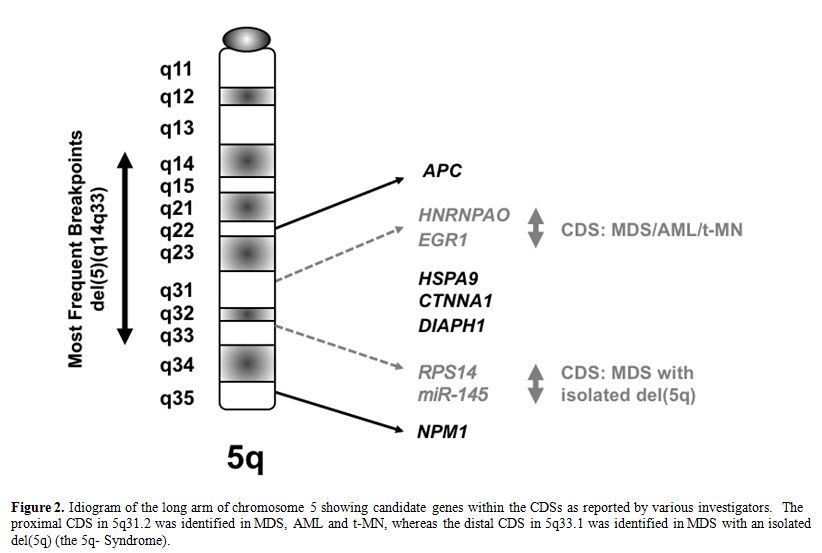
Figure 2. Idiogram of the long arm of chromosome 5 showing candidate genes within the CDSs as reported by various investigators. The proximal CDS in 5q31.2 was identified in MDS, AML and t-MN, whereas the distal CDS in 5q33.1 was identified in MDS with an isolated del(5q) (the 5q- Syndrome).
Despite intense efforts, the identification of tumor suppressor genes (TSGs) on chromosomes 5 and 7 has been challenging. Molecular analysis of the 19 candidate genes within the CDS of 5q31.2 performed in our laboratory, and by Graubert et al., did not reveal inactivating mutations in the remaining alleles, nor was there evidence of transcriptional silencing (Godley and Le Beau, unpublished data).[9,11] Similarly, molecular analysis of all 44 genes mapping to the CDS in 5q33.1 in the 5q- Syndrome did not reveal inactivating mutations.8 These observations are compatible with a haploinsufficiency model in which loss of one allele of the relevant gene(s) on 5q perturbs cell fate .[12] A number of genes located on 5q, including RPS14,[13] EGR1,[14] NPM1,[15] APC,[16] CTNNA1,[17] HSPA9,[18] and DIAPH1,[19] have been implicated in the development of myeloid disorders due to a gene dosage effect, and are reviewed briefly below. Together, these studies support a haploinsufficiency model, in which loss of a single allele of more than one gene on 5q contributes to the pathogenesis of t-MN with del(5q).
RPS14: The gene encoding RPS14, which is required for the processing of 18S pre-rRNA, is located at 5q33.1, and was identified as a candidate disease gene in the 5q- Syndrome.[13] Downregulation of RPS14 in CD34+ bone marrow cells blocks the differentiation of erythroid cells, and increases apoptosis in differentiating erythroid cells in vitro. Studies in a mouse model suggest that a TP53-dependent mechanism underlies this syndrome.20 Of interest, the ribosomal processing defect caused by haploinsufficiency of RPS14 in the 5q- Syndrome is highly analogous to the functional ribosomal defect seen in Diamond-Blackfan anemia. Other studies have shown that haploinsufficiency of two micro-RNAs (miRNAs) that are abundant in HSPCs, miR-145 and miR-146a, are encoded by sequences near the RPS14 gene, and cooperate with loss of RPS14.[21,22] The Toll-interleukin-1 receptor domain-containing adaptor protein (TIRAP) and tumor necrosis factor receptor-associated factor-6 (TRAF6) are respective targets of these miRNAs, implicating inappropriate activation of innate immune signals in the pathogenesis of the 5q- Syndrome.[21] miR-145 also targets FLI1, a gene that promotes thrombocytopoiesis.[22] Haploinsufficiency of miR-145 may account for several features of the 5q- Syndrome, including megakaryocytic dysplasia; however, neither RPS14 nor miR-145 haploinsufficiency is predicted to confer clonal dominance.
NPM1: NPM1 is involved in ribosome biogenesis and centrosome duplication, and modulates the activity of the TP53 and CDKN2A tumor suppressors. Npm1 heterozygous mice develop erythroid dysplasia with elevated mean corpuscular volume and red cell distribution width, normal red blood cell counts and hemoglobin (Hb) levels, and dysplastic megakaryocytes.[15] However, the role of NPM1 in the pathogenesis of MDS/AML is unclear, since NPM1 is not deleted in most patients with a del(5q), nor have NPM1 mutations been identified in patients with a del(5q).[23]
CTNNA1: Liu et al. showed that the -catenin gene (CTNNA1) is down-regulated in stem and progenitor cells from MDS and AML patients with a del(5q) as compared to patients lacking del(5q), or normal HSPCs.[17] CTNNA1 is suppressed due to epigenetic silencing in HL-60 cells, a myeloid leukemia cell line used as a model for del(5q) leukemia. Reinduction of CTNNA1 expression led to reduced proliferation, and an increased frequency of apoptosis, suggesting that down-regulation of -catenin in HSPCs may contribute to transformation of myeloid cells in AML patients with a del(5q).[17] However, analysis of mice with a conditional knockout of Ctnna1 in hematopoietic cells (Ctnna1+/-), revealed no defects in hematopoiesis, or predisposition to myeloid neoplasms following mutagenesis with N-ethyl-nitrosourea (ENU).[24]
HSPA9: Heat shock protein A9 (HSPA9, also known as mortalin) is a highly conserved HSP70 family member that serves as a protein chaperone. Stimulation of hematopoietic progenitor cells with erythropoietin (EPO) results in upregulation of HSPA9, which may serve as a mediator of EPO signaling. The HSPA9 gene is distal to the CDS in 5q31.2; however, one allele of this gene is deleted in the vast majority of cases. Using lentiviral-mediated knockdown in primary human hematopoietic cells and in a murine bone marrow-transplantation model, Chen et al. found that knockdown of HSPA9 in human cells significantly delayed the maturation of erythroid precursors, increased apoptosis and decreased cell cycling; myeloid and megakaryocytic precursors were not affected.[18]In the murine Hspa9-knockdown model, HSPCs, megakaryocyte/erythrocyte progenitors, erythroid precursors, and B lymphocytes were significantly reduced in number, suggesting that Hspa9 haploinsufficiency contributes to abnormal hematopoiesis. Nonetheless, additional cooperating gene mutations would be necessary for the pathogenesis of myeloid disorders and clonal dominance.
DIAPH1: The gene encoding Diaphanous-related formin, mDia1 (DIAPH1), maps to 5q31.3, between the CDSs identified in 5q31.2 and q33.1. mDia has critical roles in actin remodeling in cell division and in response to adhesive and migratory stimuli. Diaph1+/- and Diaph1-/- mice have normal hematopoiesis, but develop age-dependent myeloproliferative defects in a small percentage of mice.[19] Eisenmann et al., have proposed that mDia1 acts as a node in a tumor-suppressor network that involves multiple 5q gene products (RPS14, EGR1, CTNNA1, and possibly APC). The network has the potential to sense dynamic changes in actin assembly, providing a homeostatic mechanism that serves to balance the regulation of growth control and differentiation in HSPCs. Although intriguing, this model awaits further experimental validation.
APC: APC (adenomatous polyposis coli) is a multifunctional tumor suppressor that is involved in the initiation and progression of colorectal cancer via regulation of the WNT signaling cascade. The APC gene is located at chromosome band 5q22.2, and is deleted in >95% of patients with a del(5q),[6] raising the question of whether haploinsufficiency of APC contributes to the development of myeloid neoplasms with loss of 5q. Qian et al. employed the Cre-loxP system to inactivate Apc in hematopoietic cells in vivo.[25] Conditional inactivation of Apc in vivo dramatically increased apoptosis and enhanced cell cycle entry of HSPCs, leading to their rapid disappearance and bone marrow failure. Conditional inactivation of a single allele of Apc in mice led to the development of severe anemia with macrocytosis and monocytosis.[16] Further characterization of the erythroid lineage revealed that erythropoiesis was blocked at the early stages of differentiation. The short-term and long-term hematopoietic stem cell populations were expanded in Apc-heterozygous mice as compared to the control littermates; however, the HSPCs had a reduced capacity to regenerate hematopoiesis in vivo in the absence of a single allele of Apc. Apc heterozygous myeloid progenitor cells displayed an increased frequency of apoptosis, and decreased in vitro colony-forming capacity, recapitulating several characteristic features of myeloid neoplasms with a del(5q). These results indicated that haploinsufficiency of Apc impairs hematopoiesis, and raised the possibility that loss of function of APC contributes to the development of MDS and AML with a del(5q).
EGR1: The early growth response 1 gene (EGR1) encodes a member of the WT-1 family of transcription factors and contains 3 Cys2His2 Zn fingers that bind the GC-rich consensus sequences, GCG(G/T)GGGCG.26 In the mouse, Egr1 has been shown to be an early response gene, and mediates the cellular response to growth factors, mitogens, and stress stimuli. [26] Egr1+/- or Egr1-/- mouse embryonic fibroblasts bypass senescence and have immortalized growth characteristics, suggesting a role for Egr1 as a “gatekeeper” of p53-dependent growth regulation. EGR1 has also been shown to act as a TSG in several human tumors, including breast carcinomas and non-small cell lung cancer. [27] Recently, Egr1 has been shown to be a direct transcriptional regulator of many known TSGs, e.g., Tp53, Cdkn1a/p21, Tgfb and Pten. [27]
We characterized the hematopoietic potential of WT, Egr1+/-, and Egr1-/- mice, and found that heterozygous or homozygous loss of Egr1 alone under normal physiological conditions does not affect the hematopoietic potential of murine marrow.[14] However, Wagers and colleagues have shown that Egr1-deficient mice show spontaneous mobilization of HSPCs into the periphery, identifying Egr1 as a transcriptional regulator of stem cell migration.[28] To investigate whether loss of Egr1 cooperates with secondary mutations to induce leukemia in the mouse, we treated WT, Egr1+/-, and Egr1-/- mice with a single dose of 100mg/kg ENU at 4 weeks or 20 weeks of age. ENU was chosen because it is an alkylating agent, and may recapitulate the effects of alkylating agent chemotherapy in patients who develop t-MN. ENU-treated Egr1+/- and Egr1-/- mice developed a myeloproliferative disorder with ineffective erythropoiesis (MPD) at a high incidence. Our data suggests that loss of a single allele of Egr1 cooperates with mutations induced by an alkylating agent in the development of malignant myeloid diseases in mice. Nevertheless, Egr1 haploinsufficiency alone in vivo does not result in expansion of the HSPCs orabnormalities in adult hematopoiesis.[14]
To identify genes that cooperate with loss of Egr1 in murine leukemogenesis, we are using a forward genetic screen by retroviral insertional mutagenesis.[29] We have injected cohorts of wild type (WT) (n=61) and Egr1+/- (n=77) neonatal mice with the MOL4070LTR retrovirus. WT and Egr1+/- mice developed disease at an onset of 7 mos. of age. Egr1+/- mice develop AML or an MPD-like leukemia with a shorter latency and at a higher overall frequency than WT littermate controls (median survival: Egr1 WT 456 days, Egr1 +/- 397 days, p=0.038) (Figure 3). Of note is that the incidence of myeloid disease is higher in Egr1+/- mice (68%) than in WT mice (49%), indicating that loss of one allele of Egr1 shifts the disease spectrum from the more common lymphoid neoplasm to myeloid neoplasms. To identify cooperating cancer genes, we have cloned over 1500 retroviral integrations from myeloid neoplasms developing in 11 Egr1 WT and 19 Egr1+/- mice. Ligation-mediated polymerase chain reaction was used to identify retroviral integrations at 30 common integration sites. The most common integration site was upstream of the Evi1 gene. Of particular interest, 16/33 (48%) of Egr1+/- mice, as compared to 5/17 (29%) of Egr1 WT mice had elevated Evi1 expression in spleen cells from diseased mice, which correlated with retroviral integrations upstream of Evi1.
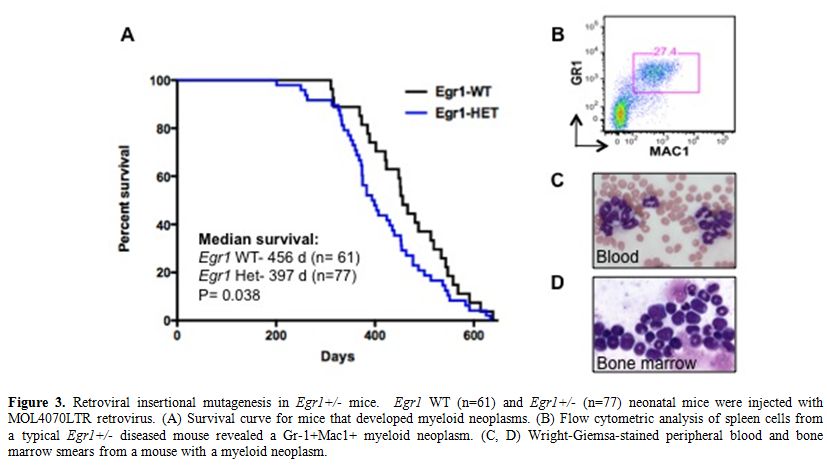
Figure 3. Retroviral insertional mutagenesis in Egr1+/- mice. Egr1 WT (n=61) and Egr1+/- (n=77) neonatal mice were injected with MOL4070LTR retrovirus. (A) Survival curve for mice that developed myeloid neoplasms. (B) Flow cytometric analysis of spleen cells from a typical Egr1+/- diseased mouse revealed a Gr-1+Mac1+ myeloid neoplasm. (C, D) Wright-Giemsa-stained peripheral blood and bone marrow smears from a mouse with a myeloid neoplasm.
EVI1 is overexpressed in AMLs, and is associated with a poor prognosis. Activation of EVI1 in hematopoietic cells can occur by juxtaposition of the gene to enhancer elements of the ribophorin gene located at 3q21 as a result of the inv(3)(q21q26.2)/t(3;3)(q21;q26.2) and, in the t(3;21)(q26.2;q22), as part of the fusion RUNX1/EVI1 mRNA transcribed from the der(3) chromosome. EVI is also overexpressed in 70% of AMLs with -7/del(7q), as well as in AMLs with translocations involving MLL at 11q23, and in many cases with a complex karyotype. [30,31]
EVI1 encodes a transcription factor that contains a seven-zinc-finger domain at the N-terminal end, a three-finger domain in the central part of the molecule, and an acidic domain distal to the second group of zinc fingers. EVI1 interacts with a number of transcriptional and epigenetic regulators (CREBBP, CTBP, HDAC, KAT2B (P/CAF), SMAD3, GATA1, GATA2, DNMT3A, and DNMT3B), and mediates chromatin modifications and DNA hypermethylation. Depending on it’s binding partners, EVI1 can act as a transcriptional activator to promote the proliferation of HSPCs, e.g., when bound to GATA2, or as a transcriptional repressor inhibiting erythroid differentiation, e.g., when bound to GATA1. In addition, EVI1 impairs myelopoiesis by de-regulation of multiple transcription factors, including RUNX1 and PU.1.[32,33] This data raises the possibility that there are unique EVI1 interactions in each cytogenetic risk group that contribute to the development of disease. Engineered over-expression of EVI1 in hematopoietic cells of transgenic mice or via bone marrow transplantation does not result in leukemia, indicating that additional cooperating genetic mutations are required for the pathogenesis of myeloid neoplasms. In the case of del(5q), haploinsufficiency of EGR1 in concert with high EVI1 may act to promote proliferation and self-renewal of HSPCs as well as to disrupt erythroid and myeloid differentiation.
HNRNPA0: A core set of genes has been identified as “master regulators” of myeloid differentiation. At the level of the granulocyte-monocyte progenitor, overall fate determination involves a balance of two opposing forces: PU.1 promotes monocytic differentiation, whereas CEBPA promotes granulocytic differentiation. Monocytic differentiation is promoted by the PU.1-induced transcription factors EGR2 and NAB2, whereas granulocytic differentiation is promoted by GFI1. EGR1/2 and NAB2 have been found to suppress the expression of GFI1 and its downstream targets; conversely, GFI1 suppresses EGR1, EGR2 and NAB2.[34] In the context of the del(5q), EGR1 haploinsufficiency would be expected to deregulate myeloid cell differentiation, favoring granulocytic over monocytic differentiation. The HNRNPA0 gene is also located within the CDS of 5q31.2, and is expressed at reduced levels in CD34+ cells from patients with MDS characterized by a del(5q) (Young and Le Beau, unpublished data). The HNRNPA0 protein is a member of the hnRNPA/B family of RNA-binding proteins, and has been shown to regulate transcript stability via binding to the AU-rich element of mRNAs. Using shRNAs in mouse hematopoietic cells, we demonstrated that knockdown of Hnrnpa0 leads to a decrease in the stability of Egr2 transcripts (Young and Le Beau, unpublished data). Thus, loss of a single allele of EGR1 and HNRNPA0 as a result of a del(5q) may lead to a synergistic disruption of EGR1/2 activity during leukemogenesis.
Alterations in Gene Function
A growing body of evidence suggests that mutations of multiple genes are involved in the pathogenesis and progression of t-MN. The involved genes fall into two main classes, namely, genes encoding hematopoietic transcription factors or proteins that regulate cytokine signaling pathways (Table 2). The RAS signaling cascade is downstream of a number of activated cytokine receptors, including the FLT3, IL3, and GM-CSF receptors; thus, this signaling pathway plays a pivotal role in hematopoiesis. Constitutively activating point mutations of NRAS, typically involving codons 12, 13, or 61, have been detected at high frequency in hematological malignancies (10-15% in t-MN).[35] Mutations of the FMS-like tyrosine kinase 3 (FLT3) gene, including both point mutations within the tyrosine kinase domain and internal tandem duplications (ITDs), are among the most common genetic changes seen in AML de novo (15-35% of cases), but are less common in t-MN (0-12%).[35,36] Mutations of NPM1 also occur frequently in AML (35% of adult cases), but are less frequent in patients with recurring cytogenetic abnormalities, and in t-MN (4-10%). Of note, the NPM1 gene located at 5q35 is not mutated in MDS with a del(5q).[15]
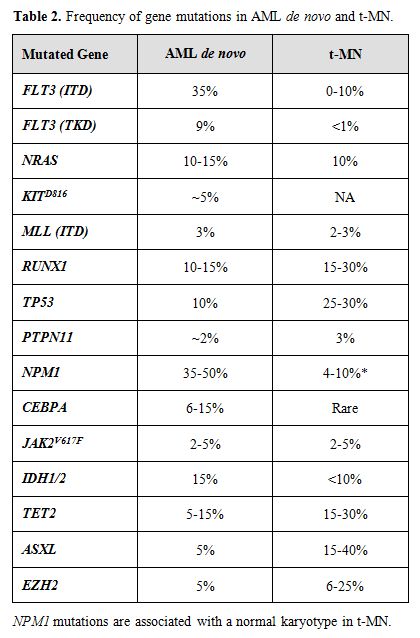
Table 2. Frequency of gene mutations in AML de novo and t-MN.
The Runt-related transcription factor 1 gene (RUNX1), also known as AML1, encodes the DNA-binding subunit of the heterodimeric core-binding factor (CBF) complex, which is essential for definitive hematopoiesis. Point mutations in the RUNX1 Runt (DNA-binding) domain have been reported in AML and MDS (10-15%), particularly in MDS secondary to atomic bomb radiation exposure or treatment. Similarly, the incidence is higher in t-MN (15-30%).[37] Moreover, RUNX1 mutations are associated with activating mutations of the RAS pathway, -7/del(7q), and a shorter overall survival.
The TP53 tumor suppressor gene encodes an essential checkpoint protein that monitors the integrity of the genome, and arrests cell cycle progression in response to DNA damage. Mutations of TP53 are observed in primary MDS and AML de novo (5-10%) and, more commonly, in t-MN (25-30%). [35,38] The spectrum of mutations includes missense mutations in exons 4-8, as well as loss of the wild type allele, typically as a result of a cytogenetic abnormality of 17p. In t-MN, TP53 mutations are associated with -5/del(5q) and a complex karyotype.
The role of epigenetic changes in the pathogenesis and treatment of MDS and AML is becoming increasingly important. Transcriptional silencing via DNA methylation of the CDKN2B (p15INK4B) gene is observed in a high percentage of patients with t-MN, and is associated with -7/del(7q), and a poor prognosis.[39,40] Other genes that may be affected by DNA methylation include the CTNNA1 gene on 5q.
Transcriptome Sequencing of t-MN
To identify expressed genetic variants that distinguish the two subtypes of t-MN, we have used next generation sequencing of the mRNA transcriptome of primary patient leukemia samples. This technology has become significantly faster and cheaper,41 and has several advantages over other systems-level genomic approaches. Although whole genome sequencing of two de novo AML leukemias identified several mutations,[42,43] mRNA sequencing enables analysis across a larger and more diverse set of samples at a fraction of the cost and time. Transcriptome sequencing better distinguishes expression levels and mRNA isoforms as compared to microarray analysis,[44,45] and enables identification of cryptic gene fusions that cannot be detected by cytogenetic analysis.[46] In addition to expression data, this method enables genotyping of the expressed coding region of the genome to identify inherited polymorphisms and somatic mutations.[47]
We hypothesized that the two t-MN subgroups, alkylating agent-related and topoisomerase II inhibitor-related, have unique, genome-wide patterns of transcriptionally expressed genetic variation that drive leukemogenesis. To test this, we employed Illumina paired-end technology to sequence the mRNA from the leukemia cells of 23 patients with t-MN (McNerney et al., unpublished data). These are comprised of 13 samples with del(5q) or -7/del(7q), or with complex karyotypes, and 12 cases with other karyotypes, including a normal karyotype or a balanced translocation. In addition, leukemia cells from13 patients with AML de novo with a spectrum of cytogenetic abnormalities comparable to the t-MN samples were similarly analyzed.
Interestingly, there are significant differences between t-MN and AML de novo samples at the level of gene expression. t-MN samples exhibit different gene expression patterns as well as variable usage of transcript isoforms. This implies that there are underlying genetic differences between t-MN and AML de novo, consistent with the fundamental biological differences between these two diseases. In addition, there is significant differential gene expression between leukemias with abnormalities of chromosomes 5 and/or 7, as compared to those with other cytogenetic patterns. By comparing RNA-sequencing data to genome wide copy number variation using single nucleotide polymorphism arrays, we are identifying those genes (genome-wide as well as those mapping to chromosomes 5 and 7) that exhibit expression level differences secondary to DNA-dosage effects.
Using the RNA-sequencing data to genotype the samples, over 13,000 single nucleotide variants (SNVs) have been identified per sample, which is similar in number to other cancer samples. Phenotypically relevant SNVs are prioritized by population-level allele frequency estimates, evolutionary constraint estimates, and the predicted coding region change. This analysis yields approximately 200 rare, predicted deleterious SNVs per sample. Some of these variants include mutations that were previously reported in AML, such as mutations involving the FLT3, TP53, RUNX1, and TET2 genes. However, mutations have not been reported previously in t-MN in most of the genes containing these variants, and many frequently occur in the same gene or pathway in multiple samples. A subset of the genes carry rare, deleterious variants, which occur preferentially in leukemias with abnormalities of chromosome 5 and/or 7. Intriguingly, the vast majority of rare, deleterious variants are inherited, which may guide us towards defining genetic predispositions to malignancy, in general, and to t-MN in particular. It is expected that genome-wide analyses across a large number of samples will result in the identification of the spectrum and frequency of expressed genetic variants in t-MN subtypes and the clinical impact of deleterious variants.
Models for the Pathogenesis of t-MN
Extensive experimental evidence indicates that more than one mutation is required for the pathogenesis of hematological malignant diseases. Moreover, these mutations cooperate to confer a proliferative and/or antiapoptotic activity, as well as impair normal differentiation pathways. Haploinsufficiency for a gene(s) on 7q and 5q is likely to be an initiating mutation. Pedersen-Bjergaard and colleagues have proposed 8 different pathways that are involved in progression to t-MN.[48] Pathway I consists of patients who have abnormalities of chromosome 7, without chromosome 5 abnormalities. These patients often present with mutations of the RAS pathway (KRAS, NRAS, NF1, PTPN11), and methylation silencing of p15 (CDKN2B), and they have a poor prognosis. Loss of TAL1, GATA1, and EKLF expression in t-MN with a -7/del(7q) may result in impaired differentiation, whereas overexpression of FLT3, PIK3C2B, and BCL2 result in a proliferative/survival advantage.49 Pathway II comprises patients with a del(5q) with or without abnormalities of chromosome 7, and a poor prognosis (Figure 4). Haploinsufficiency of multiple, cooperating genes on 5q is likely to be the initiating event. Genomic instability and complex karyotypes with gain of chromosome 8, and loss of 12p, 13q, 16q, 17p (TP53 locus), chromosome 18, and 20q, as well as mutations of TP53 are often observed in this subgroup. In t-MN patients with del(5q), loss of expression of IRF8 may lead to impaired differentiation and/or a survival advantage, whereas increased expression of cell cycle regulatory proteins (CCNA2, CCNE2, CDC2) would result in a proliferative advantage.[49]
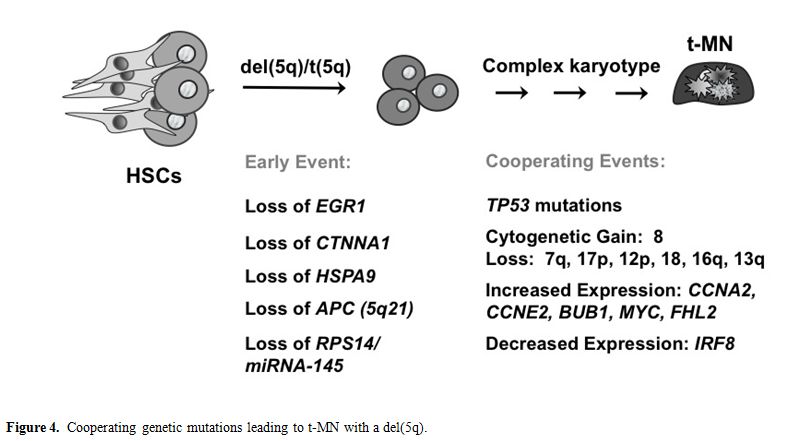
Figure 4. Cooperating genetic mutations leading to t-MN with a del(5q).
Pathway III consists of patients with translocations of 11q23. Alterations of pathway IV convey the best prognosis for patients with t-MN, and include the t(8;21) or inv(16). Pathway V comprises patients who present with therapy-related acute promyelocytic leukemia with the t(15;17) resulting in the PML-RARA fusion and a good prognosis. Pathway VI involves balanced translocations of NUP98 at 11p15. Pathway VII includes t-MN with a normal karyotype. Recently, internal tandem duplications of FLT3 and MLL, and NPM1 mutations have been described in a few of these patients. Pathway VIII includes patients with other chromosomal abnormalities. New technologies, such as high-throughput genomics technologies, will facilitate further delineation of the genetic pathways leading to t-MN.
Concluding Remarks
t-MN remains one of the most adverse complications of successful therapy for a variety of malignant and non-malignant conditions. The factors that place individual patients at risk are beginning to be elucidated, and are critical for risk-assessment, to allow individualized therapy directed at minimizing the development of this disease. Moreover, characterizing the genetic pathways that give rise to t-MN will lead to a greater understanding of genetic susceptibility to this disease, as well as the molecular features of the disease and, ultimately, may lead to more targeted therapies for its treatment.
Acknowledgements
We thank the patients, and the many members of the Leukemia Program at the University of Chicago who participated in these studies, especially Drs. Sonali M. Smith, Theodore Karrison, and Lucy A. Godley.
Supported by grants CA40046 and CA14599 from the National Cancer Institute.
t-MN is a late complication of cytotoxic therapy (radiation and/or chemotherapy) used in the treatment of both malignant and non-malignant diseases.[1-4] Several distinct cytogenetic and clinical subtypes of t-MN are recognized that are closely associated with the nature of the preceding treatment. Patients who develop t-MN following alkylating agent therapy (~75% of cases) typically show a latency of 3-7 years from alkylating agent exposure (median 5 years), insidious disease onset with an antecedent MDS with peripheral cytopenias, loss, deletion, or rearrangement of chromosomes 5 and/or 7, and a poor prognosis (median survival <8 months).[1] Typically, all three hematopoietic cell lineages (erythroid, myeloid, and megakaryocytic) are involved in the dysplastic process (trilineage dysplasia), suggesting that the disease arises in a multipotent hematopoietic stem or progenitor cell (HSPC). In contrast, patients who develop t-MN following treatment with drugs targeting topoisomerase II are younger, have a shorter latency period (2-3 years), and present with AML. Balanced translocations involving MLL at 11q23, RUNX1 at 21q22, CBFB at 16q22, or PML (15q24) and RARA (17q12) are common in this subgroup, suggesting that these cytogenetic subsets of t-MN arise in a lineage committed progenitor cell.[3-5] Survival times of t-MN patients are often short, because this disorder is less responsive to current forms of therapy than is AML de novo.[1,3,4]
t-MN represents an important model for cancer. The incidence of t-MN is rising, as a result of the increasing number of cancer survivors at risk of developing this disorder and the changes in therapeutic trends. Secondly, t-MN provides a unique opportunity to examine the effects of mutagens on carcinogenesis in humans, as well as the role of genetic susceptibility to cancer.[3] Finally, the mechanisms of leukemogenesis that are uncovered in t-MN will likely apply to those subtypes of AML de novo, which share the same cytogenetic abnormalities, e.g., AML de novo with abnormalities of chromosome 5 or 7. In this chapter, we review the genetic characteristics of t-MN with an emphasis on defining the genetic pathways leading to t-MN with a del(5q).
Cytogenetic Analyses
Table 1 summarizes the cytogenetic pattern in the recently updated University of Chicago series of 386 consecutive patients with t-MN. Of these, 349 (90.4%) had a clonal chromosomal abnormality, including 259 (67%) with a clonal abnormality leading to loss, deletion, or rearrangements of chromosomes 5 and/or 7 (referred to as del(5q)/t(5q) and -7/del(7q) herein),4 (Le Beau and Larson, unpublished data). Overall, 164 patients (42%) had abnormalities of chromosome 5, and 180 (47%) had abnormalities of chromosome 7. Of these patients, eighty-five patients had abnormalities of both chromosomes 5 and 7. A del(5q) was the most common structural abnormality. The pattern of numerical and structural abnormalities is shown in Figure 1, and illustrates that t-MN is associated with a complex karyotype, with a predominance of the loss of genetic material.
In other studies, we evaluated the cytogenetic pattern of 3,444 patients with primary MDS, AML de novo, or t-MN evaluated over the past 35 years by our Cancer Cytogenetics Laboratory. Of these, 553 (16%) patients had del(5q)/t(5q), and 597 (17.3%) had -7/del(7q) (Le Beau et al., unpublished data). Complex karyotypes were associated with abnormalities of chromosome 5, rather than chromosome 7. Recurring abnormalities observed at a high frequency (>20%) in patients with del(5q) included +8, and loss of 13q, 16q, 17p (40% of cases), chromosome 18, and 20q, which frequently occured in the same clone (Figure 1, and data not shown).

Table 1. Cytogenetic abnormalities in 386 patients with t-MN.

Figure 1. Cytogenetic abnormalities in patients with t-MN (N=386) in the University of Chicago series. Gain of chromosomal material is depicted by green bars to the right of each chromosome, and loss of chromosomal material is depicted by red bars. Numbers above the bars indicate the number of patients with this abnormality. Arrowheads identify the location of the breakpoints of structural rearrangements.
Identification of Haploinsufficient Myeloid Suppressor Genes on 5q
Several groups of investigators have defined a commonly deleted segment (CDS) on the long arm of chromosome 5 predicted to contain a myeloid tumor suppressor gene. [6-8] Using cytogenetic and molecular analysis of MDS, AML and t-MN with a del(5q), we previously defined a region of 970 kilobase within 5q31.2, flanked by D5S479 and D5S500, that is deleted in all patients (Figure 2), and determined the genomic sequence of this region.6,9 In subsequent studies, we generated a transcript map of the CDS, and identified and cloned 19 genes;9 the CDS also contains one miRNA. The functions of the proteins encoded by these genes are diverse, and include the regulation of mitosis and the G2 checkpoint, transcriptional control, and translational regulation.
MDS with an isolated del(5q) (the 5q- Syndrome) is a distinct subtype of MDS, characterized by a macrocytic anemia, female predominance, and a favorable outcome, with a low risk of transformation to AML.10 Boultwood and colleagues identified a 1.5 Mb CDS within 5q33.1 between D5S413 and GLRA1 .8 This region is distal to the CDS in 5q31.2 found in patients with AML with a del(5q). In summary, the existing data suggest that there are two non-overlapping CDSs: 5q31.2 in the more aggressive form of MDS, AML de novo, and t-MN, and 5q33.1 in the 5q- Syndrome (Figure 2).

Figure 2. Idiogram of the long arm of chromosome 5 showing candidate genes within the CDSs as reported by various investigators. The proximal CDS in 5q31.2 was identified in MDS, AML and t-MN, whereas the distal CDS in 5q33.1 was identified in MDS with an isolated del(5q) (the 5q- Syndrome).
Despite intense efforts, the identification of tumor suppressor genes (TSGs) on chromosomes 5 and 7 has been challenging. Molecular analysis of the 19 candidate genes within the CDS of 5q31.2 performed in our laboratory, and by Graubert et al., did not reveal inactivating mutations in the remaining alleles, nor was there evidence of transcriptional silencing (Godley and Le Beau, unpublished data).[9,11] Similarly, molecular analysis of all 44 genes mapping to the CDS in 5q33.1 in the 5q- Syndrome did not reveal inactivating mutations.8 These observations are compatible with a haploinsufficiency model in which loss of one allele of the relevant gene(s) on 5q perturbs cell fate .[12] A number of genes located on 5q, including RPS14,[13] EGR1,[14] NPM1,[15] APC,[16] CTNNA1,[17] HSPA9,[18] and DIAPH1,[19] have been implicated in the development of myeloid disorders due to a gene dosage effect, and are reviewed briefly below. Together, these studies support a haploinsufficiency model, in which loss of a single allele of more than one gene on 5q contributes to the pathogenesis of t-MN with del(5q).
RPS14: The gene encoding RPS14, which is required for the processing of 18S pre-rRNA, is located at 5q33.1, and was identified as a candidate disease gene in the 5q- Syndrome.[13] Downregulation of RPS14 in CD34+ bone marrow cells blocks the differentiation of erythroid cells, and increases apoptosis in differentiating erythroid cells in vitro. Studies in a mouse model suggest that a TP53-dependent mechanism underlies this syndrome.20 Of interest, the ribosomal processing defect caused by haploinsufficiency of RPS14 in the 5q- Syndrome is highly analogous to the functional ribosomal defect seen in Diamond-Blackfan anemia. Other studies have shown that haploinsufficiency of two micro-RNAs (miRNAs) that are abundant in HSPCs, miR-145 and miR-146a, are encoded by sequences near the RPS14 gene, and cooperate with loss of RPS14.[21,22] The Toll-interleukin-1 receptor domain-containing adaptor protein (TIRAP) and tumor necrosis factor receptor-associated factor-6 (TRAF6) are respective targets of these miRNAs, implicating inappropriate activation of innate immune signals in the pathogenesis of the 5q- Syndrome.[21] miR-145 also targets FLI1, a gene that promotes thrombocytopoiesis.[22] Haploinsufficiency of miR-145 may account for several features of the 5q- Syndrome, including megakaryocytic dysplasia; however, neither RPS14 nor miR-145 haploinsufficiency is predicted to confer clonal dominance.
NPM1: NPM1 is involved in ribosome biogenesis and centrosome duplication, and modulates the activity of the TP53 and CDKN2A tumor suppressors. Npm1 heterozygous mice develop erythroid dysplasia with elevated mean corpuscular volume and red cell distribution width, normal red blood cell counts and hemoglobin (Hb) levels, and dysplastic megakaryocytes.[15] However, the role of NPM1 in the pathogenesis of MDS/AML is unclear, since NPM1 is not deleted in most patients with a del(5q), nor have NPM1 mutations been identified in patients with a del(5q).[23]
CTNNA1: Liu et al. showed that the -catenin gene (CTNNA1) is down-regulated in stem and progenitor cells from MDS and AML patients with a del(5q) as compared to patients lacking del(5q), or normal HSPCs.[17] CTNNA1 is suppressed due to epigenetic silencing in HL-60 cells, a myeloid leukemia cell line used as a model for del(5q) leukemia. Reinduction of CTNNA1 expression led to reduced proliferation, and an increased frequency of apoptosis, suggesting that down-regulation of -catenin in HSPCs may contribute to transformation of myeloid cells in AML patients with a del(5q).[17] However, analysis of mice with a conditional knockout of Ctnna1 in hematopoietic cells (Ctnna1+/-), revealed no defects in hematopoiesis, or predisposition to myeloid neoplasms following mutagenesis with N-ethyl-nitrosourea (ENU).[24]
HSPA9: Heat shock protein A9 (HSPA9, also known as mortalin) is a highly conserved HSP70 family member that serves as a protein chaperone. Stimulation of hematopoietic progenitor cells with erythropoietin (EPO) results in upregulation of HSPA9, which may serve as a mediator of EPO signaling. The HSPA9 gene is distal to the CDS in 5q31.2; however, one allele of this gene is deleted in the vast majority of cases. Using lentiviral-mediated knockdown in primary human hematopoietic cells and in a murine bone marrow-transplantation model, Chen et al. found that knockdown of HSPA9 in human cells significantly delayed the maturation of erythroid precursors, increased apoptosis and decreased cell cycling; myeloid and megakaryocytic precursors were not affected.[18]In the murine Hspa9-knockdown model, HSPCs, megakaryocyte/erythrocyte progenitors, erythroid precursors, and B lymphocytes were significantly reduced in number, suggesting that Hspa9 haploinsufficiency contributes to abnormal hematopoiesis. Nonetheless, additional cooperating gene mutations would be necessary for the pathogenesis of myeloid disorders and clonal dominance.
DIAPH1: The gene encoding Diaphanous-related formin, mDia1 (DIAPH1), maps to 5q31.3, between the CDSs identified in 5q31.2 and q33.1. mDia has critical roles in actin remodeling in cell division and in response to adhesive and migratory stimuli. Diaph1+/- and Diaph1-/- mice have normal hematopoiesis, but develop age-dependent myeloproliferative defects in a small percentage of mice.[19] Eisenmann et al., have proposed that mDia1 acts as a node in a tumor-suppressor network that involves multiple 5q gene products (RPS14, EGR1, CTNNA1, and possibly APC). The network has the potential to sense dynamic changes in actin assembly, providing a homeostatic mechanism that serves to balance the regulation of growth control and differentiation in HSPCs. Although intriguing, this model awaits further experimental validation.
APC: APC (adenomatous polyposis coli) is a multifunctional tumor suppressor that is involved in the initiation and progression of colorectal cancer via regulation of the WNT signaling cascade. The APC gene is located at chromosome band 5q22.2, and is deleted in >95% of patients with a del(5q),[6] raising the question of whether haploinsufficiency of APC contributes to the development of myeloid neoplasms with loss of 5q. Qian et al. employed the Cre-loxP system to inactivate Apc in hematopoietic cells in vivo.[25] Conditional inactivation of Apc in vivo dramatically increased apoptosis and enhanced cell cycle entry of HSPCs, leading to their rapid disappearance and bone marrow failure. Conditional inactivation of a single allele of Apc in mice led to the development of severe anemia with macrocytosis and monocytosis.[16] Further characterization of the erythroid lineage revealed that erythropoiesis was blocked at the early stages of differentiation. The short-term and long-term hematopoietic stem cell populations were expanded in Apc-heterozygous mice as compared to the control littermates; however, the HSPCs had a reduced capacity to regenerate hematopoiesis in vivo in the absence of a single allele of Apc. Apc heterozygous myeloid progenitor cells displayed an increased frequency of apoptosis, and decreased in vitro colony-forming capacity, recapitulating several characteristic features of myeloid neoplasms with a del(5q). These results indicated that haploinsufficiency of Apc impairs hematopoiesis, and raised the possibility that loss of function of APC contributes to the development of MDS and AML with a del(5q).
EGR1: The early growth response 1 gene (EGR1) encodes a member of the WT-1 family of transcription factors and contains 3 Cys2His2 Zn fingers that bind the GC-rich consensus sequences, GCG(G/T)GGGCG.26 In the mouse, Egr1 has been shown to be an early response gene, and mediates the cellular response to growth factors, mitogens, and stress stimuli. [26] Egr1+/- or Egr1-/- mouse embryonic fibroblasts bypass senescence and have immortalized growth characteristics, suggesting a role for Egr1 as a “gatekeeper” of p53-dependent growth regulation. EGR1 has also been shown to act as a TSG in several human tumors, including breast carcinomas and non-small cell lung cancer. [27] Recently, Egr1 has been shown to be a direct transcriptional regulator of many known TSGs, e.g., Tp53, Cdkn1a/p21, Tgfb and Pten. [27]
We characterized the hematopoietic potential of WT, Egr1+/-, and Egr1-/- mice, and found that heterozygous or homozygous loss of Egr1 alone under normal physiological conditions does not affect the hematopoietic potential of murine marrow.[14] However, Wagers and colleagues have shown that Egr1-deficient mice show spontaneous mobilization of HSPCs into the periphery, identifying Egr1 as a transcriptional regulator of stem cell migration.[28] To investigate whether loss of Egr1 cooperates with secondary mutations to induce leukemia in the mouse, we treated WT, Egr1+/-, and Egr1-/- mice with a single dose of 100mg/kg ENU at 4 weeks or 20 weeks of age. ENU was chosen because it is an alkylating agent, and may recapitulate the effects of alkylating agent chemotherapy in patients who develop t-MN. ENU-treated Egr1+/- and Egr1-/- mice developed a myeloproliferative disorder with ineffective erythropoiesis (MPD) at a high incidence. Our data suggests that loss of a single allele of Egr1 cooperates with mutations induced by an alkylating agent in the development of malignant myeloid diseases in mice. Nevertheless, Egr1 haploinsufficiency alone in vivo does not result in expansion of the HSPCs orabnormalities in adult hematopoiesis.[14]
To identify genes that cooperate with loss of Egr1 in murine leukemogenesis, we are using a forward genetic screen by retroviral insertional mutagenesis.[29] We have injected cohorts of wild type (WT) (n=61) and Egr1+/- (n=77) neonatal mice with the MOL4070LTR retrovirus. WT and Egr1+/- mice developed disease at an onset of 7 mos. of age. Egr1+/- mice develop AML or an MPD-like leukemia with a shorter latency and at a higher overall frequency than WT littermate controls (median survival: Egr1 WT 456 days, Egr1 +/- 397 days, p=0.038) (Figure 3). Of note is that the incidence of myeloid disease is higher in Egr1+/- mice (68%) than in WT mice (49%), indicating that loss of one allele of Egr1 shifts the disease spectrum from the more common lymphoid neoplasm to myeloid neoplasms. To identify cooperating cancer genes, we have cloned over 1500 retroviral integrations from myeloid neoplasms developing in 11 Egr1 WT and 19 Egr1+/- mice. Ligation-mediated polymerase chain reaction was used to identify retroviral integrations at 30 common integration sites. The most common integration site was upstream of the Evi1 gene. Of particular interest, 16/33 (48%) of Egr1+/- mice, as compared to 5/17 (29%) of Egr1 WT mice had elevated Evi1 expression in spleen cells from diseased mice, which correlated with retroviral integrations upstream of Evi1.

Figure 3. Retroviral insertional mutagenesis in Egr1+/- mice. Egr1 WT (n=61) and Egr1+/- (n=77) neonatal mice were injected with MOL4070LTR retrovirus. (A) Survival curve for mice that developed myeloid neoplasms. (B) Flow cytometric analysis of spleen cells from a typical Egr1+/- diseased mouse revealed a Gr-1+Mac1+ myeloid neoplasm. (C, D) Wright-Giemsa-stained peripheral blood and bone marrow smears from a mouse with a myeloid neoplasm.
EVI1 is overexpressed in AMLs, and is associated with a poor prognosis. Activation of EVI1 in hematopoietic cells can occur by juxtaposition of the gene to enhancer elements of the ribophorin gene located at 3q21 as a result of the inv(3)(q21q26.2)/t(3;3)(q21;q26.2) and, in the t(3;21)(q26.2;q22), as part of the fusion RUNX1/EVI1 mRNA transcribed from the der(3) chromosome. EVI is also overexpressed in 70% of AMLs with -7/del(7q), as well as in AMLs with translocations involving MLL at 11q23, and in many cases with a complex karyotype. [30,31]
EVI1 encodes a transcription factor that contains a seven-zinc-finger domain at the N-terminal end, a three-finger domain in the central part of the molecule, and an acidic domain distal to the second group of zinc fingers. EVI1 interacts with a number of transcriptional and epigenetic regulators (CREBBP, CTBP, HDAC, KAT2B (P/CAF), SMAD3, GATA1, GATA2, DNMT3A, and DNMT3B), and mediates chromatin modifications and DNA hypermethylation. Depending on it’s binding partners, EVI1 can act as a transcriptional activator to promote the proliferation of HSPCs, e.g., when bound to GATA2, or as a transcriptional repressor inhibiting erythroid differentiation, e.g., when bound to GATA1. In addition, EVI1 impairs myelopoiesis by de-regulation of multiple transcription factors, including RUNX1 and PU.1.[32,33] This data raises the possibility that there are unique EVI1 interactions in each cytogenetic risk group that contribute to the development of disease. Engineered over-expression of EVI1 in hematopoietic cells of transgenic mice or via bone marrow transplantation does not result in leukemia, indicating that additional cooperating genetic mutations are required for the pathogenesis of myeloid neoplasms. In the case of del(5q), haploinsufficiency of EGR1 in concert with high EVI1 may act to promote proliferation and self-renewal of HSPCs as well as to disrupt erythroid and myeloid differentiation.
HNRNPA0: A core set of genes has been identified as “master regulators” of myeloid differentiation. At the level of the granulocyte-monocyte progenitor, overall fate determination involves a balance of two opposing forces: PU.1 promotes monocytic differentiation, whereas CEBPA promotes granulocytic differentiation. Monocytic differentiation is promoted by the PU.1-induced transcription factors EGR2 and NAB2, whereas granulocytic differentiation is promoted by GFI1. EGR1/2 and NAB2 have been found to suppress the expression of GFI1 and its downstream targets; conversely, GFI1 suppresses EGR1, EGR2 and NAB2.[34] In the context of the del(5q), EGR1 haploinsufficiency would be expected to deregulate myeloid cell differentiation, favoring granulocytic over monocytic differentiation. The HNRNPA0 gene is also located within the CDS of 5q31.2, and is expressed at reduced levels in CD34+ cells from patients with MDS characterized by a del(5q) (Young and Le Beau, unpublished data). The HNRNPA0 protein is a member of the hnRNPA/B family of RNA-binding proteins, and has been shown to regulate transcript stability via binding to the AU-rich element of mRNAs. Using shRNAs in mouse hematopoietic cells, we demonstrated that knockdown of Hnrnpa0 leads to a decrease in the stability of Egr2 transcripts (Young and Le Beau, unpublished data). Thus, loss of a single allele of EGR1 and HNRNPA0 as a result of a del(5q) may lead to a synergistic disruption of EGR1/2 activity during leukemogenesis.
Alterations in Gene Function
A growing body of evidence suggests that mutations of multiple genes are involved in the pathogenesis and progression of t-MN. The involved genes fall into two main classes, namely, genes encoding hematopoietic transcription factors or proteins that regulate cytokine signaling pathways (Table 2). The RAS signaling cascade is downstream of a number of activated cytokine receptors, including the FLT3, IL3, and GM-CSF receptors; thus, this signaling pathway plays a pivotal role in hematopoiesis. Constitutively activating point mutations of NRAS, typically involving codons 12, 13, or 61, have been detected at high frequency in hematological malignancies (10-15% in t-MN).[35] Mutations of the FMS-like tyrosine kinase 3 (FLT3) gene, including both point mutations within the tyrosine kinase domain and internal tandem duplications (ITDs), are among the most common genetic changes seen in AML de novo (15-35% of cases), but are less common in t-MN (0-12%).[35,36] Mutations of NPM1 also occur frequently in AML (35% of adult cases), but are less frequent in patients with recurring cytogenetic abnormalities, and in t-MN (4-10%). Of note, the NPM1 gene located at 5q35 is not mutated in MDS with a del(5q).[15]

Table 2. Frequency of gene mutations in AML de novo and t-MN.
The Runt-related transcription factor 1 gene (RUNX1), also known as AML1, encodes the DNA-binding subunit of the heterodimeric core-binding factor (CBF) complex, which is essential for definitive hematopoiesis. Point mutations in the RUNX1 Runt (DNA-binding) domain have been reported in AML and MDS (10-15%), particularly in MDS secondary to atomic bomb radiation exposure or treatment. Similarly, the incidence is higher in t-MN (15-30%).[37] Moreover, RUNX1 mutations are associated with activating mutations of the RAS pathway, -7/del(7q), and a shorter overall survival.
The TP53 tumor suppressor gene encodes an essential checkpoint protein that monitors the integrity of the genome, and arrests cell cycle progression in response to DNA damage. Mutations of TP53 are observed in primary MDS and AML de novo (5-10%) and, more commonly, in t-MN (25-30%). [35,38] The spectrum of mutations includes missense mutations in exons 4-8, as well as loss of the wild type allele, typically as a result of a cytogenetic abnormality of 17p. In t-MN, TP53 mutations are associated with -5/del(5q) and a complex karyotype.
The role of epigenetic changes in the pathogenesis and treatment of MDS and AML is becoming increasingly important. Transcriptional silencing via DNA methylation of the CDKN2B (p15INK4B) gene is observed in a high percentage of patients with t-MN, and is associated with -7/del(7q), and a poor prognosis.[39,40] Other genes that may be affected by DNA methylation include the CTNNA1 gene on 5q.
Transcriptome Sequencing of t-MN
To identify expressed genetic variants that distinguish the two subtypes of t-MN, we have used next generation sequencing of the mRNA transcriptome of primary patient leukemia samples. This technology has become significantly faster and cheaper,41 and has several advantages over other systems-level genomic approaches. Although whole genome sequencing of two de novo AML leukemias identified several mutations,[42,43] mRNA sequencing enables analysis across a larger and more diverse set of samples at a fraction of the cost and time. Transcriptome sequencing better distinguishes expression levels and mRNA isoforms as compared to microarray analysis,[44,45] and enables identification of cryptic gene fusions that cannot be detected by cytogenetic analysis.[46] In addition to expression data, this method enables genotyping of the expressed coding region of the genome to identify inherited polymorphisms and somatic mutations.[47]
We hypothesized that the two t-MN subgroups, alkylating agent-related and topoisomerase II inhibitor-related, have unique, genome-wide patterns of transcriptionally expressed genetic variation that drive leukemogenesis. To test this, we employed Illumina paired-end technology to sequence the mRNA from the leukemia cells of 23 patients with t-MN (McNerney et al., unpublished data). These are comprised of 13 samples with del(5q) or -7/del(7q), or with complex karyotypes, and 12 cases with other karyotypes, including a normal karyotype or a balanced translocation. In addition, leukemia cells from13 patients with AML de novo with a spectrum of cytogenetic abnormalities comparable to the t-MN samples were similarly analyzed.
Interestingly, there are significant differences between t-MN and AML de novo samples at the level of gene expression. t-MN samples exhibit different gene expression patterns as well as variable usage of transcript isoforms. This implies that there are underlying genetic differences between t-MN and AML de novo, consistent with the fundamental biological differences between these two diseases. In addition, there is significant differential gene expression between leukemias with abnormalities of chromosomes 5 and/or 7, as compared to those with other cytogenetic patterns. By comparing RNA-sequencing data to genome wide copy number variation using single nucleotide polymorphism arrays, we are identifying those genes (genome-wide as well as those mapping to chromosomes 5 and 7) that exhibit expression level differences secondary to DNA-dosage effects.
Using the RNA-sequencing data to genotype the samples, over 13,000 single nucleotide variants (SNVs) have been identified per sample, which is similar in number to other cancer samples. Phenotypically relevant SNVs are prioritized by population-level allele frequency estimates, evolutionary constraint estimates, and the predicted coding region change. This analysis yields approximately 200 rare, predicted deleterious SNVs per sample. Some of these variants include mutations that were previously reported in AML, such as mutations involving the FLT3, TP53, RUNX1, and TET2 genes. However, mutations have not been reported previously in t-MN in most of the genes containing these variants, and many frequently occur in the same gene or pathway in multiple samples. A subset of the genes carry rare, deleterious variants, which occur preferentially in leukemias with abnormalities of chromosome 5 and/or 7. Intriguingly, the vast majority of rare, deleterious variants are inherited, which may guide us towards defining genetic predispositions to malignancy, in general, and to t-MN in particular. It is expected that genome-wide analyses across a large number of samples will result in the identification of the spectrum and frequency of expressed genetic variants in t-MN subtypes and the clinical impact of deleterious variants.
Models for the Pathogenesis of t-MN
Extensive experimental evidence indicates that more than one mutation is required for the pathogenesis of hematological malignant diseases. Moreover, these mutations cooperate to confer a proliferative and/or antiapoptotic activity, as well as impair normal differentiation pathways. Haploinsufficiency for a gene(s) on 7q and 5q is likely to be an initiating mutation. Pedersen-Bjergaard and colleagues have proposed 8 different pathways that are involved in progression to t-MN.[48] Pathway I consists of patients who have abnormalities of chromosome 7, without chromosome 5 abnormalities. These patients often present with mutations of the RAS pathway (KRAS, NRAS, NF1, PTPN11), and methylation silencing of p15 (CDKN2B), and they have a poor prognosis. Loss of TAL1, GATA1, and EKLF expression in t-MN with a -7/del(7q) may result in impaired differentiation, whereas overexpression of FLT3, PIK3C2B, and BCL2 result in a proliferative/survival advantage.49 Pathway II comprises patients with a del(5q) with or without abnormalities of chromosome 7, and a poor prognosis (Figure 4). Haploinsufficiency of multiple, cooperating genes on 5q is likely to be the initiating event. Genomic instability and complex karyotypes with gain of chromosome 8, and loss of 12p, 13q, 16q, 17p (TP53 locus), chromosome 18, and 20q, as well as mutations of TP53 are often observed in this subgroup. In t-MN patients with del(5q), loss of expression of IRF8 may lead to impaired differentiation and/or a survival advantage, whereas increased expression of cell cycle regulatory proteins (CCNA2, CCNE2, CDC2) would result in a proliferative advantage.[49]

Figure 4. Cooperating genetic mutations leading to t-MN with a del(5q).
Pathway III consists of patients with translocations of 11q23. Alterations of pathway IV convey the best prognosis for patients with t-MN, and include the t(8;21) or inv(16). Pathway V comprises patients who present with therapy-related acute promyelocytic leukemia with the t(15;17) resulting in the PML-RARA fusion and a good prognosis. Pathway VI involves balanced translocations of NUP98 at 11p15. Pathway VII includes t-MN with a normal karyotype. Recently, internal tandem duplications of FLT3 and MLL, and NPM1 mutations have been described in a few of these patients. Pathway VIII includes patients with other chromosomal abnormalities. New technologies, such as high-throughput genomics technologies, will facilitate further delineation of the genetic pathways leading to t-MN.
Concluding Remarks
t-MN remains one of the most adverse complications of successful therapy for a variety of malignant and non-malignant conditions. The factors that place individual patients at risk are beginning to be elucidated, and are critical for risk-assessment, to allow individualized therapy directed at minimizing the development of this disease. Moreover, characterizing the genetic pathways that give rise to t-MN will lead to a greater understanding of genetic susceptibility to this disease, as well as the molecular features of the disease and, ultimately, may lead to more targeted therapies for its treatment.
Acknowledgements
We thank the patients, and the many members of the Leukemia Program at the University of Chicago who participated in these studies, especially Drs. Sonali M. Smith, Theodore Karrison, and Lucy A. Godley.
Supported by grants CA40046 and CA14599 from the National Cancer Institute.
References
- Ben-Chetrit E, Godley LA, Larson R.
Therapy-related myeloid leukemia. Semin Oncol. 2008; 35:418-29. doi:10.1053/j.seminoncol.2008.04.012
PMid:18692692 PMCid:2600445

- Vardiman JW, Thiele J, Arber DA, Brunning
RD, Borowitz MJ, Porwit A, Harris NL, Le Beau MM, Hellström-Lindberg E,
Tefferi A, Bloomfield CD. The 2008 revision of the World Health
Organization (WHO) classification of myeloid neoplasms and acute
leukemia: rationale and important changes. Blood. 2009; 114:937-51. doi:10.1182/blood-2009-03-209262PMid:19357394

- Allan JM, Travis LB. Mechanisms of
therapy-related carcinogenesis. Nat Rev Cancer. 2005;
5:943-55.doi:10.1038/nrc1749 PMid:16294218

- Smith SM, Le Beau MM, Huo D, Karrison T,
Sobecks RM, Anastasi J, Vardiman JW, Rowley JD, Larson R.
Clinical-cytogenetic associations in 306 patients with therapy-related
myelodysplasia and myeloid leukemia: the University of Chicago series.
Blood. 2003; 102:43-2.doi:10.1182/blood-2002-11-3343
PMid:12623843

- Pedersen-Bjergaard J, Philip P. Balanced
translocations involving chromosome bands 11q23 and 21q22 are highly
characteristic of myelodysplasia and leukemia following therapy with
cytostatic agents targeting at DNA-topoisomerase II. Blood. 1991;
78:1147-48. PMid:1651134

- Zhao N, Stoffel A, Wang PW, Eisenbart JD,
Espinosa R, 3rd, Larson R, Le Beau MM. Molecular delineation of the
smallest commonly deleted region of chromosome 5 in malignant myeloid
diseases to 1-1.5 Mb and preparation of a PAC-based physical map. Proc
Natl Acad Sci USA. 1997; 94:6948-53.doi:10.1073/pnas.94.13.6948

- Fairman J, Chumakov I, Chinault AC, Nowell
PC, Nagarajan L. Physical mapping of the minimal region of loss in 5q-
chromosome. Proc Natl Acad Sci USA. 1995;
92:7406-10.doi:10.1073/pnas.92.16.7406

- Boultwood J, Fidler C, Strickson AJ,
Watkins F, Gama S, Kearney L, Tosi S, Kasprzyk A, Cheng JF, Jaju RJ,
Wainscoat JS. Narrowing and genomic annotation of the commonly deleted
region of the 5q- syndrome. Blood. 2002;
99:4638-41.doi:10.1182/blood.V99.12.4638
PMid:12036901

- Lai F, Godley LA, Joslin J, Fernald AA, Liu
J, Espinosa R,
3rd, Zhao N, Pamintuan L, Till BG, Larson R, Qian Z, Le Beau MM.
Transcript map and comparative analysis of the 1.5-Mb commonly deleted
segment of human 5q31 in malignant myeloid diseases with a del(5q).
Genomics. 2001; 71:235-45.doi:10.1006/geno.2000.6414
PMid:11161817

- Hasserjian RP, Le Beau MM, List AF, Bennett JM, Thiele J. MDS with isolated del(5q). In: Swerdlow SH, Campo E, Harris NL, Haffe ES, Pileri SA, Stein H, Thiele, J, Vardiman JW, eds. WHO Classification of Tumours of Haematopoeitic and Lymphoid Tissues. Lyon, IARC. 2008; 102
- Graubert TA, M.A. Payton, Shao J, Walgren
RA, Monahan RS,
J.L. Frater JL, Walshauser MA, Martin MG, Kasai Y, Walter MJ.
Integrated genomic analysis implicates haploinsufficiency of multiple
chromosome 5q31.2 genes in de novo myelodysplastic syndromes
pathogenesis. Plos One. 2009; 4:e4583.doi:10.1371/journal.pone.0004583
PMCid:2642994

- Shannon KM, Le Beau MM. Cancer: hay in a
haystack. Nature 2008; 451:252-53. doi:10.1038/451252a PMid:18202630

- Ebert BL, Pretz J, Bosco J, Chang CY,
Tamayo P, Galili N,
Raza A, Root DE, Attar E, Ellis SR, Golub TR. Identification of RPS14
as a 5q- syndrome gene by RNA interference screen. Nature. 2008;
451:335-39.doi:10.1038/nature06494 PMid:18202658

- Joslin JM, Fernald AA, Tennant TR, Davis
EM, Kogan SC,
Anastasi J, Crispino JD, Le Beau MM, Haploinsufficiency of EGR1, a
candidate gene in the del(5q), leads to the development of myeloid
disorders. Blood. 2007; 110:719-26. doi:10.1182/blood-2007-01-068809
PMid:17420284 PMCid:1924479

- Sportoletti P, S. S. Grisendi S, Majid SM,
Cheng K,
Clohessy JG, Viale A, Teruya-Feldstein J, Pandolfi PP. Npm1 is a
haploinsufficient suppressor of myeloid and lymphoid malignancies in
the mouse. Blood. 2008; 111:3859-62.doi:10.1182/blood-2007-06-098251
PMCid:2275037

- Wang J, Fernald AA, Anastasi J, Le Beau
MM, Qian Z.
Haploinsufficiency of Apc leads to ineffective hematopoiesis. Blood.
2010;115:3481-88. doi:10.1182/blood-2009-11-251835
PMid:20065296

- Liu TX, Becker MW, Jelinek J, Wu WS, Deng
M, Mikhalkevich
N, Hsu K, Bloomfield CD, Stone RM, DeAngelo DJ, Galinsky IA, Issa JP,
Clarke MF, Look AT. Chromosome 5q deletion and epigenetic suppression
of the gene encoding alpha-catenin (CTNNA1) in myeloid cell
transformation. Nat Med. 2007; 13:78-83. doi:10.1038/nm1512
PMid:17159988

- Chen TH, Kambal A, Krysiak K, Walshauser
MA, Raju G,
Tibbitts JF, Walter MJ. Knockdown of Hspa9, a del(5q31.2) gene, results
in a decrease in hematopoietic progenitors in mice. Blood. 2011 Feb
3;117:1530-9. Epub 2010 Dec 1. PMID: 21123823

- Eisenmann KM, Dykema KJ, Matheson SF, Kent
NF, DeWard AD,
West RA, Tibes R, Furge KA, Alberts AS. 5q- myelodysplastic syndromes:
chromosome 5q genes direct a tumor suppression network sensing actin
dynamics. Oncogene. 2009; 28:3429-41. progenitors in mice. Blood. 2011;
117:1530-39

- Barlow JL, Drynan LF, Hewett DR, Holmes
LR, Lorenzo-Abalde
S, Lane AL, Jolin HE, Pannell R, Middleton AJ, Wong SH, Warren AJ,
Wainscoat JS, Boultwood J, McKenzie AN. A p53-dependent mechanism
underlies macrocytic anemia in a mouse model of human 5q- syndrome. Nat
Med. 2009;16:59-66.doi:10.1038/nm.2063
PMid:19966810 PMCid:2803774

- Starczynowski DT, Kuchenbauer F,
Argiropoulos B, Sung S,
Morin R, Muranyi A, Hirst M, Hogge D, Marra M, Wells RA, Buckstein R,
Lam W, Humphries RK, Karsan A. Identification of miR-145 and miR-146a
as mediators of the 5q- syndrome phenotype. Nat Med. 2010;
16:49-58.doi:10.1038/nm.2054 PMid:19898489

- Kumar M, Narla A, Nonami A, Ball B, Chin C, Chen C, Kutok JL, Galili N, Raza A, Attar EC, Gilliland DG, Jacks T, Ebert BL. Coordinate loss of a microRNA mir145 and a protein-coding gene RPS14 cooperate in the pathogenesis of 5q- Syndrome. Blood. 2009; 114:947
- Shiseki M, Kitagawa Y, Wang YH, Yoshinaga
K, Kondo T,
Kuroiwa H. Lack of nucleophosmin mutation in patients with
myelodysplastic syndrome and acute myeloid leukemia with chromosome 5
abnormalities. Leukaemia Lymphoma. 2007; 48:2141-44. doi:10.1080/10428190701615900
PMid:17990177

- Mikhalkevich N, and Becker MW. Alpha-catenin is dispensible for normal hematopoietic stem cell function. Blood. 2009; 114: abstract 1438
- Qian Z, Chen L, Fernald AA, Williams BO,
Le Beau MM. A
critical role for Apc in hematopoietic stem and progenitor cell
survival. J Exp Med. 2008; 205:2163-75.doi:10.1084/jem.20080578
PMid:18725524 PMCid:2526209

- Sukhatme VP, Cao XM, Chang LC, Tsai-Morris
CH, Stamenkovich
D, Ferreira PC, Cohen DR, Edwards SA, Shows TB, Curran TA, Le Beau MM,
Adamson ED. A zinc finger-encoding gene coregulated with c-fos during
growth and differentiation, and after cellular depolarization. Cell.
1988; 53:37-43. doi:10.1016/0092-8674(88)90485-0

- Baron V, Adamson ED, Calogero A, Ragona G,
Mercola D. The
transcription factor Egr1 is a direct regulator of multiple tumor
suppressors including TGFbeta1, PTEN, p53, and fibronectin. Cancer Gene
Ther. 2006; 13:115-124. doi:10.1038/sj.cgt.7700896
PMid:16138117
PMCid:2455793

- Min IM, Pietramaggiori G, Kim FF, Passegué
E, Stevenson KE,
Wagers AJ. The transcription factor EGR1 controls both the
proliferation and localization of hematopoietic stem cells. Cell Stem
Cell. 2008; 10:380-91. doi:10.1016/j.stem.2008.01.015
PMid:18397757

- Wolff L, Koller R, Hu X, Anver MR. A
Moloney murine
leukemia virus-based retrovirus with 4070A long terminal repeat
sequences induces a high incidence of myeloid as well as lymphoid
neoplasms. J Virol. 2003;
77:4965-71.doi:10.1128/JVI.77.8.4965-4971.2003
PMid:12663802
PMCid:152129

- Lugthart S, van Drunen E, van Norden Y,
van Hoven A,
Erpelinck CAJ, Valk PJM, Beverloo BH, Löwenberg B, Delwel R. High EVI1
levels predict adverse outcome in acute myeloid leukemia: prevalence of
EVI1 overexpression and chromosome 3q26 abnormalities underestimated.
Blood. 2008; 111:4329-37.doi:10.1182/blood-2007-10-119230
PMid:18272813

- Gröschel S, Lugthart S, Schlenk RF, Valk
PJM, Eiwen K,
Goudswaard C, van Putten WJL, Kayser S, Verdonck LF, Lübbert M,
Ossenkoppele G-J, Germing U, Schmidt-Wolf I, Schlegelberger B, Krauter
J, Ganser A, Döhner H, Löwenberg B, Döhner K, Delwel R. High EVI1
expression predicts outcome in younger adult patients with acute
myeloid leukemia and is associated with distinct cytogenetic
abnormalities. J Clin Onc. 2010; 28:2101-7. doi:10.1200/JCO.2009.26.0646
PMid:20308656

- Senyuk V, Sinha KK, Li D, Rinaldi CR,
Yanamandra S,
Nucifora G. Repression of RUNX1 activity by EVI1: A new role of EVI1 in
leukemogenesis. Can Res. 2007;
67:5658-66.doi:10.1158/0008-5472.CAN-06-3962
PMid:17575132

- Laricchia-Robbio L, Premanand K, Rinaldi
CR, Nucifora G.
EVI1 impairs yyelopoiesis by deregulation of PU.1 function. Can Res.
2009; 69:1633-42. doi:10.1158/0008-5472.CAN-08-2562
PMid:19208846

- Laslo P, Spooner CJ, Warmflash A, Lancki
DW, Lee HJ,
Sciammas R, Gantner BN, Dinner AR, Singh H. Multilineage
transcriptional priming and determination of alternate hematopoietic
cell fates. Cell. 2006; 126:755-66.doi:10.1016/j.cell.2006.06.052
PMid:16923394

- Side LE, Curtiss NP, Teel K, Kratz C, Wang
PW, Larson R, Le
Beau MM, Shannon KM. RAS, FLT3, and TP53 mutations in therapy-related
myeloid malignancies with abnormalities of chromosomes 5 and 7. Genes
Chromosomes Cancer. 2004; 39:217-23. doi:10.1002/gcc.10320
PMid:14732923

- Bacher U, Haferlach T, Kern W, Haferlach
C, Schnittger S. A
comparative study of molecular mutations in 381 patients with
myelodysplastic syndrome and in 4130 patients with acute myeloid
leukemia. Haematologica. 2007; 92:744-52.doi:10.3324/haematol.10869
PMid:17550846

- Chen CY, Lin LI, Tang JL, Ko BS, Tsay W,
Chou WC, Yao M, Wu
SJ, Tseng MH, Tien HF. RUNX1 gene mutation in primary myelodysplastic
syndrome--the mutation can be detected early at diagnosis or acquired
during disease progression and is associated with poor outcome. Br J
Haematol. 2007; 139:405-14.doi:10.1111/j.1365-2141.2007.06811.x
PMid:17910630

- Christiansen DH, Andersen MK,
Pedersen-Bjergaard J.
Mutations with loss of heterozygosity of p53 are common in
therapy-related myelodysplasia and acute myeloid leukemia after
exposure to alkylating agents and significantly associated with
deletion or loss of 5q, a complex karyotype, and a poor prognosis. J
Clin Oncol. 2001; 19:1405-13.PMid:11230485

- Christiansen DH, Andersen MK,
Pedersen-Bjergaard J.
Methylation of p15INK4B is common, is associated with deletion of genes
on chromosome arm 7q and predicts a poor prognosis in therapy-related
myelodysplasia and acute myeloid leukemia. Leukemia. 2003; 17:1813-19. doi:10.1038/sj.leu.2403054
PMid:12970781

- Leone G, Pagano L, Ben-Yehuda D, Voso MT.
Therapy-related
leukemia and myelodysplasia: susceptibility and incidence.
Haematologica. 2007; 92:1389-98. doi:10.3324/haematol.11034
PMid:17768113

- ten Bosch JR,Grody WW. Keeping up with the
next generation:
Massively parallel sequencing in clinical diagnostics. J Mol Diagn.
2008; 10:484-92. doi:10.2353/jmoldx.2008.080027
PMid:18832462
PMCid:2570630

- Ley TJ, Mardis ER, Ding L, Fulton B,
McLellan MD, Chen K,
Dooling D, Dunford-Shore BH, McGrath S, Hickenbotham M, Cook L, Abbott
R, Larson DE, Koboldt DC, Pohl C, Smith S, Hawkins A, Abbott S, Locke
D, Hillier LW, Miner T, Fulton L, Magrini V, Wylie T, Glasscock J,
Conyers J, Sander N, Shi X, Osborne JR, Minx P, Gordon D, Chinwalla A,
Zhao Y, Ries RE, Payton JE, Westervelt P, Tomasson MH, Watson M, Baty
J, Ivanovich J, Heath S, Shannon WD, Nagarajan R, Walter MJ, Link DC,
Graubert TA, DiPersio JF, Wilson RK. DNA sequencing of a
cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;
456:66-72. PMid:18987736 PMCid:2603574

- Mardis ER, Ding L, Dooling DJ, Larson DE,
McLellan MD, Chen
K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD, Fulton LA, Locke
DP, Magrini VJ, Abbott RM, Vickery TL, Reed JS, Robinson JS, Wylie T,
Smith SM, Carmichael L, Eldred JM, Harris CC, Walker J, Peck JB, Du F,
Dukes AF, Sanderson GE, Brummett AM, Clark E, McMichael JF, Meyer RJ,
Schindler JK, Pohl CS, Wallis JW, Shi X, Lin L, Schmidt H, Tang Y,
Haipek C, Wiechert ME, Ivy JV, Kalicki J, Elliott G, Ries RE, Payton
JE, Westervelt P, Tomasson MH, Watson MA, Baty J, Heath S, Shannon WD,
Nagarajan R, Link DC, Walter MJ, Graubert TA, Dipersio JF, Wilson RK,
Ley TJ. Recurring mutations found by sequencing an acute myeloid
leukemia genome. N Engl J Med. 2009;
361:1058-66.doi:10.1056/NEJMoa0903840
PMid:19657110

- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe
BJ. Deep surveying
of alternative splicing complexity in the human transcriptome by
high-throughput sequencing. Nat Genet. 2008;
40:1413-5.doi:10.1038/ng.259 PMid:18978789

- Marioni JC, Mason CE, Mane SM, Stephens M,
Gilad Y.
RNA-seq: an assessment of technical reproducibility and comparison with
gene expression arrays. Genome Res. 2008;
18:1509-17.doi:10.1101/gr.079558.108
PMid:18550803 PMCid:2527709

- Zhao Q, Caballero OL, Levy S, Stevenson
BJ, Iseli C, de
Souza SJ, Galante PA, Busam D, Leversha MA, Chadalavada K, Rogers YH,
Venter JC, Simpson AJ, Strausberg RL. Transcriptome-guided
characterization of genomic rearrangements in a breast cancer cell
line. Proc Natl Acad Sci USA. 2009; 106:1886-91. doi:10.1073/pnas.0812945106
PMid:19181860 PMCid:2633215

- Shah SP, Kobel M, Senz J, Morin RD, Clarke
BA, Wiegand KC,
Leung G, Zayed A, Mehl E, Kalloger SE, Sun M, Giuliany R, Yorida E,
Jones S, Varhol R, Swenerton KD, Miller D, Clement PB, Crane C, Madore
J, Provencher D, Leung P, DeFazio A, Khattra J, Turashvili G, Zhao Y,
Zeng T, Glover JN, Vanderhyden B, Zhao C, Parkinson CA, Jimenez-Linan
M, Bowtell DD, Mes-Masson AM, Brenton JD, Aparicio SA, Boyd N, Hirst M,
Gilks CB, Marra M, Huntsman DG. Mutation of FOXL2 in granulosa-cell
tumors of the ovary. N Engl J Med. 2009; 360:2719-29. doi:10.1056/NEJMoa0902542
PMid:19516027

- Pedersen-Bjergaard J, Andersen MK,
Christiansen DH, Nerlov
C. Genetic pathways in therapy-related myelodysplasia and acute myeloid
leukemia. Blood. 2002; 99:1909-12.doi:10.1182/blood.V99.6.1909
PMid:11877259

- Qian Z, Fernald AA, Godley LA, Larson R,
Le Beau MM.
Expression profiling of CD34+ hematopoietic stem/ progenitor cells
reveals distinct subtypes of therapy-related acute myeloid leukemia.
Proc Natl Acad Sci USA. 2002; 99:14925-30.doi:10.1073/pnas.222491799
PMid:12417757 PMCid:137521
