The Spectrum of Genetic Defects in Chronic Lymphocytic Leukemia
Davide Rossi1, Marco Fangazio1 and Gianluca Gaidano1
1 Division of Hematology, Department of Translational Medicine, Amedeo Avogadro University of Eastern Piedmont, Novara, Italy
Correspondence
to:
Davide Rossi, M.D., Ph.D., Division of Hematology, Department of
Translational Medicine, Amedeo Avogadro University of Eastern Piedmont,
Via Solaroli 17, 28100 Novara, Italy; Tel: +39-0321-660698; Fax:
+39-0321-620421; E-mail rossidav@med.unipmn.it
Published: November 13, 2012
Received: October 26, 2012
Accepted: November 9, 2012
Meditter J Hematol Infect Dis 2012, 4(1): e2012076, DOI 10.4084/MJHID.2012.076
This article is available on PDF format at:

This is an Open Access article
distributed under the terms of the
Creative Commons Attribution License
(http://creativecommons.org/licenses/by/2.0),
which permits
unrestricted use, distribution, and reproduction in any medium,
provided the original work is properly cited
Abstract
Chronic
lymphocytic leukemia (CLL) is the most common leukemia in the Western
world and shows a remarkable heterogeneity in the clinical course.
Understand the genetic basis of CLL may help in clarifying the
molecular bases of this clinical heterogeneity. Recurrent chromosomal
aberrations at 13q14, 12q, 11q22-q23 and 17p13, and TP53 mutations are
the first genetic lesions identified as drivers of the disease. While
some of these lesions are associated with poor outcome (17p13 deletion,
TP53 mutations and, to a lesser extent, 11q22-q23 deletion) others are
linked to a favorable course (13q14 deletion as sole aberration).
Recently, next generation sequencing has revealed additional recurrent
alterations in CLL targeting the NOTCH1, SF3B1, and BIRC3 genes.
NOTCH1, SF3B1, and BIRC3 lesions provide: i) new insights on the
mechanisms of leukemogenesis, tumor progression and chemoresistance in
this leukemia; ii) new biomarkers for the identification of poor risk
patients, having individually shown correlations with survival in CLL;
and iii) new therapeutic targets, especially in the setting of high
risk disease. This review will summarize the most important genetic
aberrations in CLL and how our improved knowledge of the genome of
leukemic cells may translate into improved patients' management.
Introduction
In Western countries, chronic lymphocytic leukemia (CLL) is the most frequent mature B-cell malignancy.[1,2] The course CLL ranges from very indolent, with a nearly normal life expectancy, to rapidly progressive leading to early death.[3-8] Understand the genetic basis of CLL may help in clarifying the molecular determinants of this clinical heterogeneity and improve patients' prognostication.
Recurrent chromosomal aberrations at 13q14, 12q, 11q22-q23 and 17p13 are the first genetic lesions identified as drivers of the disease, and has enabled the construction of a hierarchical model of cytogenetic abnormalities that correlates with outcome.[9] Cytogenetic lesions, however, may not entirely explain the genetic basis of CLL clinical heterogeneity, as documented by the contribution of TP53 mutation assessment in identifying high risk patients.[9] The recent major improvements in massive parallel sequencing technologies have provided an opportunity to examine the CLL genome, allowing for the identification of genomic alterations underlying the disease and for the discovery of new therapeutic targets and clinically predictive biomarkers such as NOTCH1, SF3B1 and BIRC3.[10-16]
Prevalence of Genetic Lesions at Different CLL Clinical Phases
During its history, CLL may proceed through distinct clinical phases, ranging from a pre-malignant condition known as monoclonal B-cell lymphocytosis (MBL), to overt CLL, and even transformation into an aggressive lymphoma (Richter syndrome).[1,2]
Similarly to other pre-malignant conditions, also MBL frequently harbor genetic changes that can be found in the overt disease. In MBL, 13q14 deletion occurs at the same prevalence as in overt CLL (~40-50% of cases), even when the number of circulating monoclonal CLL-like cells is extremely small, thus indicating that this lesion occurs early during the natural history of the disease.[17-21] What distinguishes MBL from CLL is the rate of occurrence of genetic lesions that are considered secondary events and that associate with poor outcome in this leukemia.[19,21] In clinical MBL, 11q22-q23 deletion, 17p13 deletion and mutations of BIRC3, TP53, NOTCH1 and SF3B1 may be observed in ~1-3% of cases, a prevalence that is significantly lower than that of CLL (Table 1).[17,19,21,22] High risk cytogenetic abnormalities have been occasionally described also in low count MBL, but the biological implications of this observation are currently unknown.[18,20]
When CLL is overt, three major clinical phases can be envisaged, including: i) newly diagnosed CLL; ii) progressive CLL; and iii) relapsed and fludarabine-refractory CLL (Table 1).[2] TP53 abnormalities, including mutations and 17p13 deletions, are observed in ~5-10% newly diagnosed CLL, in ~10% progressive CLL requiring first treatment,[9,23-32] and in ~40-50% relapsed and fludarabine-refractory CLL,[33-35] thus representing the most frequent lesions in this high risk clinical condition. Deletion of 11q22-q23 occurs in 10-15% in newly diagnosed CLL,[9,36] while its prevalence raises to 20-25% at the time of first treatment and 25-30%% at fludarabine-refractoriness.[24,29,33,34] Mutations of ATM, which is included in the minimal common region of deletion on 11q22-q23, have been shown to be present in 12% of newly diagnosed patients and in 15% progressive CLL requiring first treatment.[37-40] By combining mutations and deletions, genetic lesions of ATM occur in 25% of diagnostic samples of CLL and in 37% cases requiring first treatment.[37-40] These frequencies make ATM alterations the most common genetic lesions predicting poor outcome at CLL presentation and treatment requirement.
Among the novel genetic alterations disclosed by whole genome/exome sequencing, NOTCH1, SF3B1 and BIRC3 lesions follows the same distribution across CLL clinical phases as TP53 and ATM abnormalities (Table 1). NOTCH1 mutations recur in ~10% unselected newly diagnosed CLL while their prevalence increases to 15-20% in progressive and relapsed cases.[10,11,14] SF3B1 mutations have been identified in ~7% unselected newly diagnosed CLL, while their prevalence rises to 17% in relapsed and fludarabine-refractory patients.[12,13,16] BIRC3 lesions occur at low rate (4% of cases) in unselected newly diagnosed CLL, while are enriched among relapsed and fludarabine-refractory CLL (24% of cases).[15] Because of their recent identification and the lack of information from large clinical trials, the precise rate of occurrence of NOTCH1, BIRC3, and SF3B1 lesions at the time of first treatment requirement still remains to be clarified.
Within the spectrum of the various aspects of CLL, Richter syndrome (RS) is the most aggressive clinical phenotype because of the combined effect of chemoresistance and rapid disease kinetics. The clinical behavior of RS is strongly related to its genetic background (Table 1). The high rate of TP53 abnormalities, which occur in ~60% cases and represent the most frequent genetic lesion at the time of transformation, accounts for the chemoresistance that is very common in RS.[41] NOTCH1 mutations are the second most frequent genetic lesion in RS, where they occur in ~30% of cases.[10] Among the other high risk genetic lesions, ATM abnormalities, BIRC3 genetic lesions and SF3B1 mutations that are otherwise enriched at the time of chemorefractoriness are rare or absent in RS, thus strengthening the notion that RS is molecularly distinct from chemorefractory progression without transformation.[13,14,41]
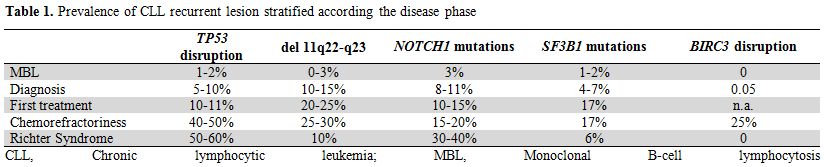
Table 1. Prevalence of CLL recurrent lesion stratified according the disease phase
TP53 Abnormalities
The tumor suppressor gene TP53 codes for a central regulator of the DNA-damage-response pathway, and its activation leads to cell-cycle arrest, DNA repair, apoptosis, or senescence through both transcription-dependent and transcriptional-independent activities.[42] Among CLL harboring TP53 abnormalities, mutations of TP53 co-occurred with deletion of the corresponding locus in ~70% of cases, consistent with a dual hit mechanism of inactivation.[43] The remaining ~30% of cases have 17p13 deletion in the absence of TP53 mutations (~20%), or TP53 mutations in the absence of 17p13 deletion (~10%). TP53 mutations are mainly represented by missense substitutions targeting the DNA-binding domain, while the remaining are truncating lesions. Mutations either directly disrupt the DNA binding domain of TP53 or cause conformational changes of the TP53 protein, thus leading to severely impaired TP53 function.[43,44]
The clinical importance of TP53 abnormalities in CLL is tightly linked to their close association with poor outcome and refractoriness, as documented by a number of observational studies and prospective trials led in both the chemotherapy and immuno-chemotherapy era. Among unselected newly diagnosed CLL, patients harboring 17p13 deletion have an estimated median overall survival (OS) of only 3-5 years.[9,45] However, it is important to stress that there is a small subgroup of patients with 17p13 deletion (and mostly mutated immunoglobulin genes) who may exhibit stable disease for years without treatment indications.[45]
The outcome of patients with 17p13 deletion and need for treatment is very poor. With the most effective regimen available today for CLL, i.e. FCR (fludarabine-cyclophosphamide-rituximab), patients with 17p13 deletion have a poor response (5% of complete response vs ~50% in non 17p13 deleted CLL), a short progression free survival (PFS) (11.2 months vs 51.8 months) and OS (38.1% at 36 months).[29] This is in line with the established importance of the wild-type TP53 protein in mediating the cytotoxicity of DNA-damaging agents including purine analogs.
A number of prospective studies suggest that, in addition to 17p13 deletion, also TP53 mutations, even in the absence of 17p13 deletion, predict poor outcome in CLL. In the GCLLSG CLL4 trial (fludarabine vs fludarabine-cyclophosphamide) no complete response were observed in TP53 mutated CLL, and the median PFS (23.3 vs 62.2 months) and OS (29.2 vs 84.6 months) were significantly shorter in the group with TP53 mutation.[30] In the GCLLSG CLL8 trial (fludarabine-cyclophosphamide vs FCR), patients with TP53 mutations showed the lowest complete response and overall response rates (6.9% vs. 36.4% and 62.1% vs. 95.3%), translating into shorter PFS (12.4 months vs. 45 months) and OS (39.3 months vs not reached in all other patients).[44] In the UK LRF CLL4 trial (chlorambucil vs fludarabine vs fludarabine-cyclophosphamide), the complete response rate of TP53 mutated patients was only 5% with a 5-years PFS of 5% and a 5-years OS of 20%.[31]
Based on these data, 17p13 deletion is the sole cytogenetic abnormality that is recommended to be tested by FISH in CLL patients requiring treatment.[2] Since CLL with TP53 mutations experience poor prognosis regardless of the presence of 17p13 deletion, the TP53 mutation analysis should be integrated into the evaluation of CLL patients before treatment initiation.[44] CLL patients carrying TP53 alterations, regardless of whether mutated or deleted, should be redirected to different therapeutic regimens compared to the standard chemo/chemoimmuno-therapies.[2,33,35,44,46]
NOTCH1 Mutations
The NOTCH1 gene encodes a heterodimeric transmembrane protein that functions as a ligand-activated transcription factor with a high conserved pathway.[47] When the NOTCH1 receptor interacts with its ligands through the extracellular subunit, two consecutive proteolytic cleavages of the protein are initiated and lead to pathway activation.[47,48] The S2 cleavage in the heterodimerization domain is performed by ADAM10, and is followed by the S3 cleavage by the γ-secretase complex. Upon activation the cleaved intracellular portion of NOTCH1 (ICN) translocates into the nucleus where it modifies the expression of target genes, including the MYC oncogene. As a transcriptional factor, NOTCH1 plays an important role in a number of cellular functions during embryogenesis and in self-renewing tissues of the adult organism, including maintenance of stem cells, cell fate specification, proliferation, and apoptosis.[48] One of the mechanisms of the NOTCH1 signal suppression is operated through the PEST [proline (P), glutamic acid (E), serine (S), and threonine (T) rich] domain that directs the activated NOTCH1 towards proteosomal degradation.[47] A major role of NOTCH1 in lymphoid cells in the adult organism is the commitment of hematopoietic progenitors to differentiate toward T lineage.[49] Conversely, in mature B-lymphocytes, NOTCH1 signaling promotes terminal differentiation to antibody-secreting cells.[50]
NOTCH1 mutations were the first molecular lesion identified through massive parallel next generation sequencing in CLL by two independent groups.[10,11] NOTCH1 mutations are significantly more frequent in CLL with unmutated, rather than mutated, immunoglobulin genes, are significantly enriched in CLL harboring trisomy 12, and identify a distinct clinico-molecular subgroup of CLL with deregulated cell cycle and short survival.[10-12,14,16,51-53]
NOTCH1 mutations in CLL mainly clusters within a hotspot in exon 34, and are commonly represented by a single 2-bp deletion (c.7544_7545delCT) that accounts for ~80-95% of all NOTCH1 mutations in this leukemia (Figure 1).[10-12,14,16,51-53] The predicted functional consequence of NOTCH1 mutations in CLL is the disruption of the C-terminal PEST domain resulting in activated NOTCH1 protein, impaired degradation and accumulation, and sustaining deregulated signaling.[11] Consistent with this notion, a number of cellular pathways are specifically altered in CLL harboring NOTCH1 mutations.[11,52]
Beside their pathogenetic role, NOTCH1 mutations also represent a new biomarker for the identification of poor risk CLL patients. NOTCH1 mutated patients have a rapidly progressive disease and a significantly shorter survival probability (21-45% at 10 years) compared to NOTCH1 wild type cases (56-66% at 10 years).[10,11,14] The poor prognosis associated with NOTCH1 mutations in CLL may be explained, at least in part, by a substantial risk (~40-50%) of developing Richter syndrome.[10,11,14]
NOTCH1 is a potential therapeutic target in CLL. Treatment with γ-secretase inhibitors induces apoptosis of CLL cells by inhibiting the enzymatic S3 cleavage necessary for NOTCH1 activation.[47,54,55] However, the limitations due to toxicity of γ-secretase inhibitors in the clinical setting suggest that alternative strategies may be needed for the therapeutic targeting of NOTCH1.
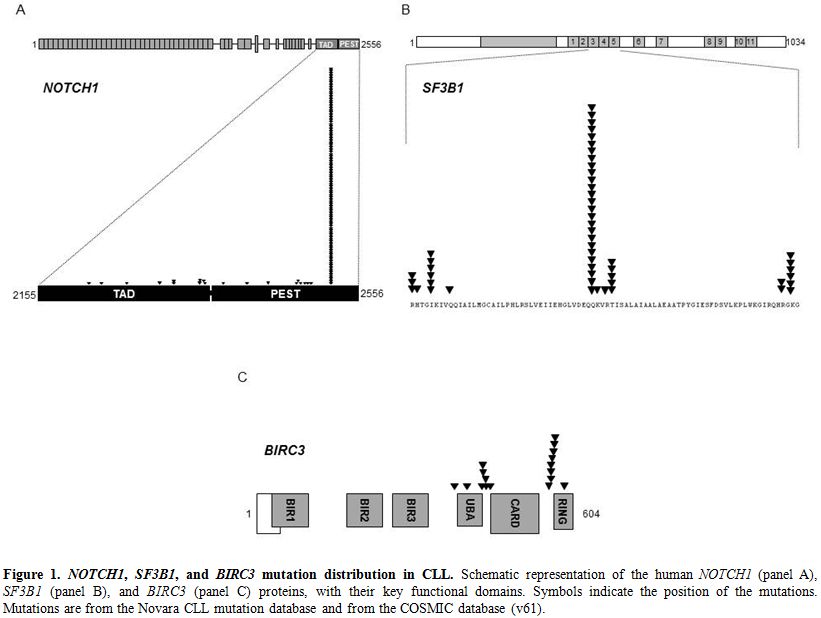
Figure 1. NOTCH1, SF3B1, and BIRC3 mutation distribution in CLL. Schematic representation of the human NOTCH1 (panel A), SF3B1 (panel B), and BIRC3 (panel C) proteins, with their key functional domains. Symbols indicate the position of the mutations. Mutations are from the Novara CLL mutation database and from the COSMIC database (v61).
SF3B1 Mutations
The spliceosome machinery, a complex of five small nuclear ribonucleoproteins (snRNPs), contributes to the formation of mature mRNA through the removal of introns in the precursor messenger RNA (pre-mRNA) of protein-encoding genes, and is involved in both normal and alternative splicing.[56] Alternative splicing can generate numerous transcript variants from a single gene, contributing to genomic complexity and potentially to cancer.[57]
SF3B1 is a core component of the U2 snRNP that recognizes the 3′ splice site at the intron-exon junctions. [56,58-61] Structurally, the SF3B1 protein has two well-defined regions: i) the N-terminal amino acid region which contains several protein-binding motifs and functions as a scaffold to facilitate its interaction with other splicing factors; and ii) the C-terminal region which contains 22 non-identical tandem repeats of the HEAT motif that meander around the SF3b complex.[56,58-61]
Whole genome/exome sequencing technologies allowed for the identification of SF3B1 as a recurrently mutated gene in CLL.[12,13,16] SF3B1 mutations in CLL cluster in selected HEAT repeats of the SF3B1 protein, target a number of hotspots (codons 662, 666, 700, 742), and are generally represented by missense substitutions (Figure 1).[12,13,16] Notably, an identical spectrum of SF3B1 mutations has been identified in other hematopoietic tumors of the myeloid compartment.[62]
The precise biological consequences of SF3B1 mutations in CLL are currently unknown. However, the clustering of SF3B1 mutations within the HEAT domains suggests that they are selected to modify SF3B1 interactions with other proteins of the spliceosome complex, thus resulting in deregulated normal and alternative mRNA splicing.[12,16]
Consistent with their accumulation in the more advanced phases of the disease, SF3B1 mutated patients show a significantly shorter overall survival (34-48% at 10 years) compared to wild type cases (60-73% at 10-years).[12,13,16]
BIRC3 Abnormalities
In CLL, activation of the NF-κB pathway contributes to the acquisition of a chemorefractory clinical phenotype and correlates with poor outcome.[63-67] The Baculoviral IAP repeat containing 3 (BIRC3) gene is one of the components of a protein complex that negatively regulates the MAP3K14 serin-threonine kinase, the downstream activator of non-canonical NF-κB signaling.[63-66]
BIRC3 was found to be recurrently disrupted by mutations, deletions, or a combination of mutations and deletions in CLL patients.[15] BIRC3 inactivating mutations and a fraction of BIRC3 deletions cause a truncation of the C-terminal RING domain of the BIRC3 protein, essential for ubiquitination, and the following proteasome degradation, of MAP3K14, and drives constitutive non-canonical NF-κB activation (Figure 1).[15]
The BIRC3 gene maps to 11q22.2, approximately 6Mb centromeric to the ATM locus. The identification of BIRC3 involvement in CLL might be important for elucidating the molecular genetics of 11q22-q23 deletion, a frequent cytogenetic abnormality predictive of poor outcome. In fact, although ATM has been regarded as the relevant gene of this chromosomal abnormality, biallelic inactivation of ATM does not exceed ~30% of cases with 11q22-q23 deletion.[36-39] The presence of an additional tumor suppressor in the 11q22-q23 region has been postulated,[40] and BIRC3 implicates a suitable candidate.
From a clinical standpoint, BIRC3 lesions contribute to clinical aggressiveness and fludarabine refractoriness in CLL.[15] Indeed, BIRC3 lesions identify a subgroup of CLL displaying poor survival (median 3.1 years) similar to that associated with TP53 abnormalities.[15]
In CLL, fludarabine refractoriness may be explained by TP53 disruption in ~40% of patients, while ~60% high risk CLL do not present TP53 abnormalities.[34] Intriguingly the distribution of BIRC3 disruption and TP53 abnormalities is mutually exclusive and BIRC3 abnormalities can recapitulate the genetics of ~40% chemorefractory and TP53 wild type CLL.
On these bases, BIRC3 disruption may contribute to expand the panel of biomarkers for the early identification of chemorefractory cases.[15] In addition, BIRC3 abnormalities provide a molecular rationale for targeting NF-κB in poor risk and chemorefractory CLL. NF-κB inhibitors are under development in CLL and pre-clinical findings suggest that these compounds might be active against chemoresistant CLL clones.[67,68]
In Western countries, chronic lymphocytic leukemia (CLL) is the most frequent mature B-cell malignancy.[1,2] The course CLL ranges from very indolent, with a nearly normal life expectancy, to rapidly progressive leading to early death.[3-8] Understand the genetic basis of CLL may help in clarifying the molecular determinants of this clinical heterogeneity and improve patients' prognostication.
Recurrent chromosomal aberrations at 13q14, 12q, 11q22-q23 and 17p13 are the first genetic lesions identified as drivers of the disease, and has enabled the construction of a hierarchical model of cytogenetic abnormalities that correlates with outcome.[9] Cytogenetic lesions, however, may not entirely explain the genetic basis of CLL clinical heterogeneity, as documented by the contribution of TP53 mutation assessment in identifying high risk patients.[9] The recent major improvements in massive parallel sequencing technologies have provided an opportunity to examine the CLL genome, allowing for the identification of genomic alterations underlying the disease and for the discovery of new therapeutic targets and clinically predictive biomarkers such as NOTCH1, SF3B1 and BIRC3.[10-16]
Prevalence of Genetic Lesions at Different CLL Clinical Phases
During its history, CLL may proceed through distinct clinical phases, ranging from a pre-malignant condition known as monoclonal B-cell lymphocytosis (MBL), to overt CLL, and even transformation into an aggressive lymphoma (Richter syndrome).[1,2]
Similarly to other pre-malignant conditions, also MBL frequently harbor genetic changes that can be found in the overt disease. In MBL, 13q14 deletion occurs at the same prevalence as in overt CLL (~40-50% of cases), even when the number of circulating monoclonal CLL-like cells is extremely small, thus indicating that this lesion occurs early during the natural history of the disease.[17-21] What distinguishes MBL from CLL is the rate of occurrence of genetic lesions that are considered secondary events and that associate with poor outcome in this leukemia.[19,21] In clinical MBL, 11q22-q23 deletion, 17p13 deletion and mutations of BIRC3, TP53, NOTCH1 and SF3B1 may be observed in ~1-3% of cases, a prevalence that is significantly lower than that of CLL (Table 1).[17,19,21,22] High risk cytogenetic abnormalities have been occasionally described also in low count MBL, but the biological implications of this observation are currently unknown.[18,20]
When CLL is overt, three major clinical phases can be envisaged, including: i) newly diagnosed CLL; ii) progressive CLL; and iii) relapsed and fludarabine-refractory CLL (Table 1).[2] TP53 abnormalities, including mutations and 17p13 deletions, are observed in ~5-10% newly diagnosed CLL, in ~10% progressive CLL requiring first treatment,[9,23-32] and in ~40-50% relapsed and fludarabine-refractory CLL,[33-35] thus representing the most frequent lesions in this high risk clinical condition. Deletion of 11q22-q23 occurs in 10-15% in newly diagnosed CLL,[9,36] while its prevalence raises to 20-25% at the time of first treatment and 25-30%% at fludarabine-refractoriness.[24,29,33,34] Mutations of ATM, which is included in the minimal common region of deletion on 11q22-q23, have been shown to be present in 12% of newly diagnosed patients and in 15% progressive CLL requiring first treatment.[37-40] By combining mutations and deletions, genetic lesions of ATM occur in 25% of diagnostic samples of CLL and in 37% cases requiring first treatment.[37-40] These frequencies make ATM alterations the most common genetic lesions predicting poor outcome at CLL presentation and treatment requirement.
Among the novel genetic alterations disclosed by whole genome/exome sequencing, NOTCH1, SF3B1 and BIRC3 lesions follows the same distribution across CLL clinical phases as TP53 and ATM abnormalities (Table 1). NOTCH1 mutations recur in ~10% unselected newly diagnosed CLL while their prevalence increases to 15-20% in progressive and relapsed cases.[10,11,14] SF3B1 mutations have been identified in ~7% unselected newly diagnosed CLL, while their prevalence rises to 17% in relapsed and fludarabine-refractory patients.[12,13,16] BIRC3 lesions occur at low rate (4% of cases) in unselected newly diagnosed CLL, while are enriched among relapsed and fludarabine-refractory CLL (24% of cases).[15] Because of their recent identification and the lack of information from large clinical trials, the precise rate of occurrence of NOTCH1, BIRC3, and SF3B1 lesions at the time of first treatment requirement still remains to be clarified.
Within the spectrum of the various aspects of CLL, Richter syndrome (RS) is the most aggressive clinical phenotype because of the combined effect of chemoresistance and rapid disease kinetics. The clinical behavior of RS is strongly related to its genetic background (Table 1). The high rate of TP53 abnormalities, which occur in ~60% cases and represent the most frequent genetic lesion at the time of transformation, accounts for the chemoresistance that is very common in RS.[41] NOTCH1 mutations are the second most frequent genetic lesion in RS, where they occur in ~30% of cases.[10] Among the other high risk genetic lesions, ATM abnormalities, BIRC3 genetic lesions and SF3B1 mutations that are otherwise enriched at the time of chemorefractoriness are rare or absent in RS, thus strengthening the notion that RS is molecularly distinct from chemorefractory progression without transformation.[13,14,41]

Table 1. Prevalence of CLL recurrent lesion stratified according the disease phase
TP53 Abnormalities
The tumor suppressor gene TP53 codes for a central regulator of the DNA-damage-response pathway, and its activation leads to cell-cycle arrest, DNA repair, apoptosis, or senescence through both transcription-dependent and transcriptional-independent activities.[42] Among CLL harboring TP53 abnormalities, mutations of TP53 co-occurred with deletion of the corresponding locus in ~70% of cases, consistent with a dual hit mechanism of inactivation.[43] The remaining ~30% of cases have 17p13 deletion in the absence of TP53 mutations (~20%), or TP53 mutations in the absence of 17p13 deletion (~10%). TP53 mutations are mainly represented by missense substitutions targeting the DNA-binding domain, while the remaining are truncating lesions. Mutations either directly disrupt the DNA binding domain of TP53 or cause conformational changes of the TP53 protein, thus leading to severely impaired TP53 function.[43,44]
The clinical importance of TP53 abnormalities in CLL is tightly linked to their close association with poor outcome and refractoriness, as documented by a number of observational studies and prospective trials led in both the chemotherapy and immuno-chemotherapy era. Among unselected newly diagnosed CLL, patients harboring 17p13 deletion have an estimated median overall survival (OS) of only 3-5 years.[9,45] However, it is important to stress that there is a small subgroup of patients with 17p13 deletion (and mostly mutated immunoglobulin genes) who may exhibit stable disease for years without treatment indications.[45]
The outcome of patients with 17p13 deletion and need for treatment is very poor. With the most effective regimen available today for CLL, i.e. FCR (fludarabine-cyclophosphamide-rituximab), patients with 17p13 deletion have a poor response (5% of complete response vs ~50% in non 17p13 deleted CLL), a short progression free survival (PFS) (11.2 months vs 51.8 months) and OS (38.1% at 36 months).[29] This is in line with the established importance of the wild-type TP53 protein in mediating the cytotoxicity of DNA-damaging agents including purine analogs.
A number of prospective studies suggest that, in addition to 17p13 deletion, also TP53 mutations, even in the absence of 17p13 deletion, predict poor outcome in CLL. In the GCLLSG CLL4 trial (fludarabine vs fludarabine-cyclophosphamide) no complete response were observed in TP53 mutated CLL, and the median PFS (23.3 vs 62.2 months) and OS (29.2 vs 84.6 months) were significantly shorter in the group with TP53 mutation.[30] In the GCLLSG CLL8 trial (fludarabine-cyclophosphamide vs FCR), patients with TP53 mutations showed the lowest complete response and overall response rates (6.9% vs. 36.4% and 62.1% vs. 95.3%), translating into shorter PFS (12.4 months vs. 45 months) and OS (39.3 months vs not reached in all other patients).[44] In the UK LRF CLL4 trial (chlorambucil vs fludarabine vs fludarabine-cyclophosphamide), the complete response rate of TP53 mutated patients was only 5% with a 5-years PFS of 5% and a 5-years OS of 20%.[31]
Based on these data, 17p13 deletion is the sole cytogenetic abnormality that is recommended to be tested by FISH in CLL patients requiring treatment.[2] Since CLL with TP53 mutations experience poor prognosis regardless of the presence of 17p13 deletion, the TP53 mutation analysis should be integrated into the evaluation of CLL patients before treatment initiation.[44] CLL patients carrying TP53 alterations, regardless of whether mutated or deleted, should be redirected to different therapeutic regimens compared to the standard chemo/chemoimmuno-therapies.[2,33,35,44,46]
NOTCH1 Mutations
The NOTCH1 gene encodes a heterodimeric transmembrane protein that functions as a ligand-activated transcription factor with a high conserved pathway.[47] When the NOTCH1 receptor interacts with its ligands through the extracellular subunit, two consecutive proteolytic cleavages of the protein are initiated and lead to pathway activation.[47,48] The S2 cleavage in the heterodimerization domain is performed by ADAM10, and is followed by the S3 cleavage by the γ-secretase complex. Upon activation the cleaved intracellular portion of NOTCH1 (ICN) translocates into the nucleus where it modifies the expression of target genes, including the MYC oncogene. As a transcriptional factor, NOTCH1 plays an important role in a number of cellular functions during embryogenesis and in self-renewing tissues of the adult organism, including maintenance of stem cells, cell fate specification, proliferation, and apoptosis.[48] One of the mechanisms of the NOTCH1 signal suppression is operated through the PEST [proline (P), glutamic acid (E), serine (S), and threonine (T) rich] domain that directs the activated NOTCH1 towards proteosomal degradation.[47] A major role of NOTCH1 in lymphoid cells in the adult organism is the commitment of hematopoietic progenitors to differentiate toward T lineage.[49] Conversely, in mature B-lymphocytes, NOTCH1 signaling promotes terminal differentiation to antibody-secreting cells.[50]
NOTCH1 mutations were the first molecular lesion identified through massive parallel next generation sequencing in CLL by two independent groups.[10,11] NOTCH1 mutations are significantly more frequent in CLL with unmutated, rather than mutated, immunoglobulin genes, are significantly enriched in CLL harboring trisomy 12, and identify a distinct clinico-molecular subgroup of CLL with deregulated cell cycle and short survival.[10-12,14,16,51-53]
NOTCH1 mutations in CLL mainly clusters within a hotspot in exon 34, and are commonly represented by a single 2-bp deletion (c.7544_7545delCT) that accounts for ~80-95% of all NOTCH1 mutations in this leukemia (Figure 1).[10-12,14,16,51-53] The predicted functional consequence of NOTCH1 mutations in CLL is the disruption of the C-terminal PEST domain resulting in activated NOTCH1 protein, impaired degradation and accumulation, and sustaining deregulated signaling.[11] Consistent with this notion, a number of cellular pathways are specifically altered in CLL harboring NOTCH1 mutations.[11,52]
Beside their pathogenetic role, NOTCH1 mutations also represent a new biomarker for the identification of poor risk CLL patients. NOTCH1 mutated patients have a rapidly progressive disease and a significantly shorter survival probability (21-45% at 10 years) compared to NOTCH1 wild type cases (56-66% at 10 years).[10,11,14] The poor prognosis associated with NOTCH1 mutations in CLL may be explained, at least in part, by a substantial risk (~40-50%) of developing Richter syndrome.[10,11,14]
NOTCH1 is a potential therapeutic target in CLL. Treatment with γ-secretase inhibitors induces apoptosis of CLL cells by inhibiting the enzymatic S3 cleavage necessary for NOTCH1 activation.[47,54,55] However, the limitations due to toxicity of γ-secretase inhibitors in the clinical setting suggest that alternative strategies may be needed for the therapeutic targeting of NOTCH1.

Figure 1. NOTCH1, SF3B1, and BIRC3 mutation distribution in CLL. Schematic representation of the human NOTCH1 (panel A), SF3B1 (panel B), and BIRC3 (panel C) proteins, with their key functional domains. Symbols indicate the position of the mutations. Mutations are from the Novara CLL mutation database and from the COSMIC database (v61).
SF3B1 Mutations
The spliceosome machinery, a complex of five small nuclear ribonucleoproteins (snRNPs), contributes to the formation of mature mRNA through the removal of introns in the precursor messenger RNA (pre-mRNA) of protein-encoding genes, and is involved in both normal and alternative splicing.[56] Alternative splicing can generate numerous transcript variants from a single gene, contributing to genomic complexity and potentially to cancer.[57]
SF3B1 is a core component of the U2 snRNP that recognizes the 3′ splice site at the intron-exon junctions. [56,58-61] Structurally, the SF3B1 protein has two well-defined regions: i) the N-terminal amino acid region which contains several protein-binding motifs and functions as a scaffold to facilitate its interaction with other splicing factors; and ii) the C-terminal region which contains 22 non-identical tandem repeats of the HEAT motif that meander around the SF3b complex.[56,58-61]
Whole genome/exome sequencing technologies allowed for the identification of SF3B1 as a recurrently mutated gene in CLL.[12,13,16] SF3B1 mutations in CLL cluster in selected HEAT repeats of the SF3B1 protein, target a number of hotspots (codons 662, 666, 700, 742), and are generally represented by missense substitutions (Figure 1).[12,13,16] Notably, an identical spectrum of SF3B1 mutations has been identified in other hematopoietic tumors of the myeloid compartment.[62]
The precise biological consequences of SF3B1 mutations in CLL are currently unknown. However, the clustering of SF3B1 mutations within the HEAT domains suggests that they are selected to modify SF3B1 interactions with other proteins of the spliceosome complex, thus resulting in deregulated normal and alternative mRNA splicing.[12,16]
Consistent with their accumulation in the more advanced phases of the disease, SF3B1 mutated patients show a significantly shorter overall survival (34-48% at 10 years) compared to wild type cases (60-73% at 10-years).[12,13,16]
BIRC3 Abnormalities
In CLL, activation of the NF-κB pathway contributes to the acquisition of a chemorefractory clinical phenotype and correlates with poor outcome.[63-67] The Baculoviral IAP repeat containing 3 (BIRC3) gene is one of the components of a protein complex that negatively regulates the MAP3K14 serin-threonine kinase, the downstream activator of non-canonical NF-κB signaling.[63-66]
BIRC3 was found to be recurrently disrupted by mutations, deletions, or a combination of mutations and deletions in CLL patients.[15] BIRC3 inactivating mutations and a fraction of BIRC3 deletions cause a truncation of the C-terminal RING domain of the BIRC3 protein, essential for ubiquitination, and the following proteasome degradation, of MAP3K14, and drives constitutive non-canonical NF-κB activation (Figure 1).[15]
The BIRC3 gene maps to 11q22.2, approximately 6Mb centromeric to the ATM locus. The identification of BIRC3 involvement in CLL might be important for elucidating the molecular genetics of 11q22-q23 deletion, a frequent cytogenetic abnormality predictive of poor outcome. In fact, although ATM has been regarded as the relevant gene of this chromosomal abnormality, biallelic inactivation of ATM does not exceed ~30% of cases with 11q22-q23 deletion.[36-39] The presence of an additional tumor suppressor in the 11q22-q23 region has been postulated,[40] and BIRC3 implicates a suitable candidate.
From a clinical standpoint, BIRC3 lesions contribute to clinical aggressiveness and fludarabine refractoriness in CLL.[15] Indeed, BIRC3 lesions identify a subgroup of CLL displaying poor survival (median 3.1 years) similar to that associated with TP53 abnormalities.[15]
In CLL, fludarabine refractoriness may be explained by TP53 disruption in ~40% of patients, while ~60% high risk CLL do not present TP53 abnormalities.[34] Intriguingly the distribution of BIRC3 disruption and TP53 abnormalities is mutually exclusive and BIRC3 abnormalities can recapitulate the genetics of ~40% chemorefractory and TP53 wild type CLL.
On these bases, BIRC3 disruption may contribute to expand the panel of biomarkers for the early identification of chemorefractory cases.[15] In addition, BIRC3 abnormalities provide a molecular rationale for targeting NF-κB in poor risk and chemorefractory CLL. NF-κB inhibitors are under development in CLL and pre-clinical findings suggest that these compounds might be active against chemoresistant CLL clones.[67,68]
References
- Swerdlow, S.H., Campo, E., Harris, N.L.,
Jaffe, E.S., Pileri, S.A., Stein, H., Thiele, J., Vardiman, J.W WHO
Classification of Tumours of Haematopoietic and Lymphoid Tissues,
Fourth Edition. (Lyon, France, 2008).
- Hallek, M., Cheson, B.D., Catovsky, D.,
Caligaris-Cappio, F., Dighiero, G., Dohner, H., Hillmen, P., Keating,
M.J., Montserrat, E., Rai, K.R., Kipps, T.J. and International Workshop
on Chronic Lymphocytic, L., Guidelines for the diagnosis and treatment
of chronic lymphocytic leukemia: a report from the International
Workshop on Chronic Lymphocytic Leukemia updating the National Cancer
Institute-Working Group 1996 guidelines. Blood 2008 111: 5446-5456. http://dx.doi.org/10.1182/blood-2007-06-093906 PMid:18216293 PMCid:2972576

- Damle, R.N., Wasil, T., Fais, F., Ghiotto,
F., Valetto, A., Allen, S.L., Buchbinder, A., Budman, D., Dittmar, K.,
Kolitz, J., Lichtman, S.M., Schulman, P., Vinciguerra, V.P., Rai, K.R.,
Ferrarini, M. and Chiorazzi, N., Ig V gene mutation status and CD38
expression as novel prognostic indicators in chronic lymphocytic
leukemia. Blood 1999 94: 1840-1847. PMid:10477712

- Hamblin, T.J., Davis, Z., Gardiner, A.,
Oscier, D.G. and Stevenson, F.K., Unmutated Ig V(H) genes are
associated with a more aggressive form of chronic lymphocytic leukemia.
Blood 1999 94: 1848-1854. PMid:10477713

- Oscier, D.G., Gardiner, A.C., Mould, S.J.,
Glide, S., Davis, Z.A., Ibbotson, R.E., Corcoran, M.M., Chapman, R.M.,
Thomas, P.W., Copplestone, J.A., Orchard, J.A. and Hamblin, T.J.,
Multivariate analysis of prognostic factors in CLL: clinical stage,
IGVH gene mutational status, and loss or mutation of the p53 gene are
independent prognostic factors. Blood 2002 100: 1177-1184.
PMid:12149195

- Crespo, M., Bosch, F., Villamor, N.,
Bellosillo, B., Colomer, D., Rozman, M., Marce, S., Lopez-Guillermo,
A., Campo, E. and Montserrat, E., ZAP-70 expression as a surrogate for
immunoglobulin-variable-region mutations in chronic lymphocytic
leukemia. N Engl J Med 2003 348: 1764-1775. http://dx.doi.org/10.1056/NEJMoa023143 PMid:12724482

- Vasconcelos, Y., Davi, F., Levy, V.,
Oppezzo, P., Magnac, C., Michel, A., Yamamoto, M., Pritsch, O.,
Merle-Beral, H., Maloum, K., Ajchenbaum-Cymbalista, F. and Dighiero,
G., Binet's staging system and VH genes are independent but
complementary prognostic indicators in chronic lymphocytic leukemia. J
Clin Oncol 2003 21: 3928-3932. http://dx.doi.org/10.1200/JCO.2003.02.134 PMid:14581416

- Rassenti, L.Z., Huynh, L., Toy, T.L., Chen,
L., Keating, M.J., Gribben, J.G., Neuberg, D.S., Flinn, I.W., Rai,
K.R., Byrd, J.C., Kay, N.E., Greaves, A., Weiss, A. and Kipps, T.J.,
ZAP-70 compared with immunoglobulin heavy-chain gene mutation status as
a predictor of disease progression in chronic lymphocytic leukemia. N
Engl J Med 2004 351: 893-901. http://dx.doi.org/10.1056/NEJMoa040857 PMid:15329427

- D’hner, H., Stilgenbauer, S., Benner, A.,
Leupolt, E., Krober, A., Bullinger, L., Dohner, K., Bentz, M. and
Lichter, P., Genomic aberrations and survival in chronic lymphocytic
leukemia. N Engl J Med 2000 343: 1910-1916. http://dx.doi.org/10.1056/NEJM200012283432602 PMid:11136261

- Fabbri, G., Rasi, S., Rossi, D., Trifonov,
V., Khiabanian, H., Ma, J., Grunn, A., Fangazio, M., Capello, D.,
Monti, S., Cresta, S., Gargiulo, E., Forconi, F., Guarini, A., Arcaini,
L., Paulli, M., Laurenti, L., Larocca, L.M., Marasca, R., Gattei, V.,
Oscier, D., Bertoni, F., Mullighan, C.G., Foa, R., Pasqualucci, L.,
Rabadan, R., Dalla-Favera, R. and Gaidano, G., Analysis of the chronic
lymphocytic leukemia coding genome: role of NOTCH1 mutational
activation. J Exp Med 2011 208: 1389-1401. http://dx.doi.org/10.1084/jem.20110921 PMid:21670202 PMCid:3135373

- Puente, X.S., Pinyol, M., Quesada, V.,
Conde, L., Ordonez, G.R., Villamor, N., Escaramis, G., Jares, P., Bea,
S., Gonzalez-Diaz, M., Bassaganyas, L., Baumann, T., Juan, M.,
Lopez-Guerra, M., Colomer, D., Tubio, J.M., Lopez, C., Navarro, A.,
Tornador, C., Aymerich, M., Rozman, M., Hernandez, J.M., Puente, D.A.,
Freije, J.M., Velasco, G., Gutierrez-Fernandez, A., Costa, D., Carrio,
A., Guijarro, S., Enjuanes, A., Hernandez, L., Yague, J., Nicolas, P.,
Romeo-Casabona, C.M., Himmelbauer, H., Castillo, E., Dohm, J.C., de
Sanjose, S., Piris, M.A., de Alava, E., San Miguel, J., Royo, R.,
Gelpi, J.L., Torrents, D., Orozco, M., Pisano, D.G., Valencia, A.,
Guigo, R., Bayes, M., Heath, S., Gut, M., Klatt, P., Marshall, J.,
Raine, K., Stebbings, L.A., Futreal, P.A., Stratton, M.R., Campbell,
P.J., Gut, I., Lopez-Guillermo, A., Estivill, X., Montserrat, E.,
Lopez-Otin, C. and Campo, E., Whole-genome sequencing identifies
recurrent mutations in chronic lymphocytic leukaemia. Nature 2011 475:
101-105. http://dx.doi.org/10.1038/nature10113 PMid:21642962 PMCid:3322590

- Quesada, V., Conde, L., Villamor, N.,
Ordonez, G.R., Jares, P., Bassaganyas, L., Ramsay, A.J., Bea, S.,
Pinyol, M., Martinez-Trillos, A., Lopez-Guerra, M., Colomer, D.,
Navarro, A., Baumann, T., Aymerich, M., Rozman, M., Delgado, J., Gine,
E., Hernandez, J.M., Gonzalez-Diaz, M., Puente, D.A., Velasco, G.,
Freije, J.M., Tubio, J.M., Royo, R., Gelpi, J.L., Orozco, M., Pisano,
D.G., Zamora, J., Vazquez, M., Valencia, A., Himmelbauer, H., Bayes,
M., Heath, S., Gut, M., Gut, I., Estivill, X., Lopez-Guillermo, A.,
Puente, X.S., Campo, E. and Lopez-Otin, C., Exome sequencing identifies
recurrent mutations of the splicing factor SF3B1 gene in chronic
lymphocytic leukemia. Nat Genet 2012 44: 47-52. http://dx.doi.org/10.1038/ng.1032 PMid:22158541

- Rossi, D., Bruscaggin, A., Spina, V.,
Rasi, S., Khiabanian, H., Messina, M., Fangazio, M., Vaisitti, T.,
Monti, S., Chiaretti, S., Guarini, A., Del Giudice, I., Cerri, M.,
Cresta, S., Deambrogi, C., Gargiulo, E., Gattei, V., Forconi, F.,
Bertoni, F., Deaglio, S., Rabadan, R., Pasqualucci, L., Foa, R.,
Dalla-Favera, R. and Gaidano, G., Mutations of the SF3B1 splicing
factor in chronic lymphocytic leukemia: association with progression
and fludarabine-refractoriness. Blood 2011 118: 6904-6908. http://dx.doi.org/10.1182/blood-2011-08-373159 PMid:22039264

- Rossi, D., Rasi, S., Fabbri, G., Spina,
V., Fangazio, M., Forconi, F., Marasca, R., Laurenti, L., Bruscaggin,
A., Cerri, M., Monti, S., Cresta, S., Fama, R., De Paoli, L., Bulian,
P., Gattei, V., Guarini, A., Deaglio, S., Capello, D., Rabadan, R.,
Pasqualucci, L., Dalla-Favera, R., Foa, R. and Gaidano, G., Mutations
of NOTCH1 are an independent predictor of survival in chronic
lymphocytic leukemia. Blood 2012 119: 521-529. http://dx.doi.org/10.1182/blood-2011-09-379966 PMid:22077063

- Rossi, D., Fangazio, M., Rasi, S.,
Vaisitti, T., Monti, S., Cresta, S., Chiaretti, S., Del Giudice, I.,
Fabbri, G., Bruscaggin, A., Spina, V., Deambrogi, C., Marinelli, M.,
Fama, R., Greco, M., Daniele, G., Forconi, F., Gattei, V., Bertoni, F.,
Deaglio, S., Pasqualucci, L., Guarini, A., Dalla-Favera, R., Foa, R.
and Gaidano, G., Disruption of BIRC3 associates with fludarabine
chemorefractoriness in TP53 wild-type chronic lymphocytic leukemia.
Blood 2012 119: 2854-2862. http://dx.doi.org/10.1182/blood-2011-12-395673 PMid:22308293

- Wang, L., Lawrence, M.S., Wan, Y.,
Stojanov, P., Sougnez, C., Stevenson, K., Werner, L., Sivachenko, A.,
DeLuca, D.S., Zhang, L., Zhang, W., Vartanov, A.R., Fernandes, S.M.,
Goldstein, N.R., Folco, E.G., Cibulskis, K., Tesar, B., Sievers, Q.L.,
Shefler, E., Gabriel, S., Hacohen, N., Reed, R., Meyerson, M., Golub,
T.R., Lander, E.S., Neuberg, D., Brown, J.R., Getz, G. and Wu, C.J.,
SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N
Engl J Med 2011 365: 2497-2506. http://dx.doi.org/10.1056/NEJMoa1109016 PMid:22150006

- Rawstron, A.C., Bennett, F.L., O'Connor,
S.J., Kwok, M., Fenton, J.A., Plummer, M., de Tute, R., Owen, R.G.,
Richards, S.J., Jack, A.S. and Hillmen, P., Monoclonal B-cell
lymphocytosis and chronic lymphocytic leukemia. N Engl J Med 2008 359:
575-583. http://dx.doi.org/10.1056/NEJMoa075290 PMid:18687638

- Nieto, W.G., Almeida, J., Romero, A.,
Teodosio, C., Lopez, A., Henriques, A.F., Sanchez, M.L., Jara-Acevedo,
M., Rasillo, A., Gonzalez, M., Fernandez-Navarro, P., Vega, T., Orfao,
A. and Primary Health Care Group of Salamanca for the Study of, M.B.L.,
Increased frequency (12%) of circulating chronic lymphocytic
leukemia-like B-cell clones in healthy subjects using a highly
sensitive multicolor flow cytometry approach. Blood 2009 114: 33-37. http://dx.doi.org/10.1182/blood-2009-01-197368 PMid:19420353

- Rossi, D., Sozzi, E., Puma, A., De Paoli,
L., Rasi, S., Spina, V., Gozzetti, A., Tassi, M., Cencini, E.,
Raspadori, D., Pinto, V., Bertoni, F., Gattei, V., Lauria, F., Gaidano,
G. and Forconi, F., The prognosis of clinical monoclonal B cell
lymphocytosis differs from prognosis of Rai 0 chronic lymphocytic
leukaemia and is recapitulated by biological risk factors. Br J
Haematol 2009 146: 64-75. http://dx.doi.org/10.1111/j.1365-2141.2009.07711.x PMid:19438485

- Fazi, C., Scarfo, L., Pecciarini, L.,
Cottini, F., Dagklis, A., Janus, A., Talarico, A., Scielzo, C., Sala,
C., Toniolo, D., Caligaris-Cappio, F. and Ghia, P., General population
low-count CLL-like MBL persists over time without clinical progression,
although carrying the same cytogenetic abnormalities of CLL. Blood 2011
118: 6618-6625. http://dx.doi.org/10.1182/blood-2011-05-357251 PMid:21876118

- Kern, W., Bacher, U., Haferlach, C.,
Dicker, F., Alpermann, T., Schnittger, S. and Haferlach, T., Monoclonal
B-cell lymphocytosis is closely related to chronic lymphocytic
leukaemia and may be better classified as early-stage CLL. Br J
Haematol 2012 http://dx.doi.org/10.1111/j.1365-2141.2011.09010.x PMid:22224978

- Greco, M., Capello, D., Bruscaggin, A.,
Spina, V., Rasi, S., Monti, S., Ciardullo, C., Cresta, S., Fangazio,
M., Gaidano, G., Foa, R. and Rossi, D., Analysis of SF3B1 mutations in
monoclonal B-cell lymphocytosis. Hematol Oncol 2012 http://dx.doi.org/10.1002/hon.2013 PMid:22461140

- D’hner, H., Fischer, K., Bentz, M.,
Hansen, K., Benner, A., Cabot, G., Diehl, D., Schlenk, R., Coy, J.,
Stilgenbauer, S. and et al., p53 gene deletion predicts for poor
survival and non-response to therapy with purine analogs in chronic
B-cell leukemias. Blood 1995 85: 1580-1589.

- Catovsky, D., Richards, S., Matutes, E.,
Oscier, D., Dyer, M.J., Bezares, R.F., Pettitt, A.R., Hamblin, T.,
Milligan, D.W., Child, J.A., Hamilton, M.S., Dearden, C.E., Smith,
A.G., Bosanquet, A.G., Davis, Z., Brito-Babapulle, V., Else, M., Wade,
R., Hillmen, P., Group, U.K.N.C.R.I.H.O.C.S. and Group, N.C.L.L.W.,
Assessment of fludarabine plus cyclophosphamide for patients with
chronic lymphocytic leukaemia (the LRF CLL4 Trial): a randomised
controlled trial. Lancet 2007 370: 230-239. http://dx.doi.org/10.1016/S0140-6736(07)61125-8 PMid:18689542

- Zenz, T., Krober, A., Scherer, K., Habe,
S., Buhler, A., Benner, A., Denzel, T., Winkler, D., Edelmann, J.,
Schwanen, C., Dohner, H. and Stilgenbauer, S., Monoallelic TP53
inactivation is associated with poor prognosis in chronic lymphocytic
leukemia: results from a detailed genetic characterization with
long-term follow-up. Blood 2008 112: 3322-3329. http://dx.doi.org/10.1182/blood-2008-04-154070 PMid:18843282

- Dicker, F., Herholz, H., Schnittger, S.,
Nakao, A., Patten, N., Wu, L., Kern, W., Haferlach, T. and Haferlach,
C., The detection of TP53 mutations in chronic lymphocytic leukemia
independently predicts rapid disease progression and is highly
correlated with a complex aberrant karyotype. Leukemia 2009 23:
117-124. http://dx.doi.org/10.1038/leu.2008.274 PMid:19850740

- Malcikova, J., Smardova, J., Rocnova, L.,
Tichy, B., Kuglik, P., Vranova, V., Cejkova, S., Svitakova, M.,
Skuhrova Francova, H., Brychtova, Y., Doubek, M., Brejcha, M.,
Klabusay, M., Mayer, J., Pospisilova, S. and Trbusek, M., Monoallelic
and biallelic inactivation of TP53 gene in chronic lymphocytic
leukemia: selection, impact on survival, and response to DNA damage.
Blood 2009 114: 5307-5314. http://dx.doi.org/10.1182/blood-2009-07-234708 PMid:19188171

- Rossi, D., Cerri, M., Deambrogi, C.,
Sozzi, E., Cresta, S., Rasi, S., De Paoli, L., Spina, V., Gattei, V.,
Capello, D., Forconi, F., Lauria, F. and Gaidano, G., The prognostic
value of TP53 mutations in chronic lymphocytic leukemia is independent
of Del17p13: implications for overall survival and chemorefractoriness.
Clinl Cancer Res 2009 15: 995-1004. http://dx.doi.org/10.1158/1078-0432.CCR-08-1630

- Hallek, M., Fischer, K., Fingerle-Rowson,
G., Fink, A.M., Busch, R., Mayer, J., Hensel, M., Hopfinger, G., Hess,
G., von Grunhagen, U., Bergmann, M., Catalano, J., Zinzani, P.L.,
Caligaris-Cappio, F., Seymour, J.F., Berrebi, A., Jager, U., Cazin, B.,
Trneny, M., Westermann, A., Wendtner, C.M., Eichhorst, B.F., Staib, P.,
Buhler, A., Winkler, D., Zenz, T., Bottcher, S., Ritgen, M., Mendila,
M., Kneba, M., Dohner, H., Stilgenbauer, S., International Group of, I.
and German Chronic Lymphocytic Leukaemia Study, G., Addition of
rituximab to fludarabine and cyclophosphamide in patients with chronic
lymphocytic leukaemia: a randomised, open-label, phase 3 trial. Lancet
2010 376: 1164-1174. http://dx.doi.org/10.1016/S0140-6736(10)61381-5 PMid:20697090

- Zenz, T., Eichhorst, B., Busch, R.,
Denzel, T., Habe, S., Winkler, D., Buhler, A., Edelmann, J., Bergmann,
M., Hopfinger, G., Hensel, M., Hallek, M., Dohner, H. and Stilgenbauer,
S., TP53 mutation and survival in chronic lymphocytic leukemia. J Clin
Oncol 2010 28: 4473-4479. http://dx.doi.org/10.1200/JCO.2009.27.8762 PMid:21483000

- Gonzalez, D., Martinez, P., Wade, R.,
Hockley, S., Oscier, D., Matutes, E., Dearden, C.E., Richards, S.M.,
Catovsky, D. and Morgan, G.J., Mutational status of the TP53 gene as a
predictor of response and survival in patients with chronic lymphocytic
leukemia: results from the LRF CLL4 trial. J Clin Oncol 2011 29:
2223-2229. http://dx.doi.org/10.1200/JCO.2010.32.0838 PMid:20870288

- Zainuddin, N., Murray, F., Kanduri, M.,
Gunnarsson, R., Smedby, K.E., Enblad, G., Jurlander, J., Juliusson, G.
and Rosenquist, R., TP53 Mutations are infrequent in newly diagnosed
chronic lymphocytic leukemia. Leuk Res 2011 35: 272-274. http://dx.doi.org/10.1016/j.leukres.2010.08.023 PMid:19597025

- Stilgenbauer, S., Zenz, T., Winkler, D.,
Buhler, A., Schlenk, R.F., Groner, S., Busch, R., Hensel, M., Duhrsen,
U., Finke, J., Dreger, P., Jager, U., Lengfelder, E., Hohloch, K.,
Soling, U., Schlag, R., Kneba, M., Hallek, M., Dohner, H. and German
Chronic Lymphocytic Leukemia Study, G., Subcutaneous alemtuzumab in
fludarabine-refractory chronic lymphocytic leukemia: clinical results
and prognostic marker analyses from the CLL2H study of the German
Chronic Lymphocytic Leukemia Study Group. J Clin Oncol 2009 27:
3994-4001. http://dx.doi.org/10.1200/JCO.2008.21.1128 PMid:19643983

- Zenz, T., Habe, S., Denzel, T., Mohr, J.,
Winkler, D., Buhler, A., Sarno, A., Groner, S., Mertens, D., Busch, R.,
Hallek, M., Dohner, H. and Stilgenbauer, S., Detailed analysis of p53
pathway defects in fludarabine-refractory chronic lymphocytic leukemia
(CLL): dissecting the contribution of 17p deletion, TP53 mutation,
p53-p21 dysfunction, and miR34a in a prospective clinical trial. Blood
2009 114: 2589-2597. http://dx.doi.org/10.1182/blood-2009-05-224071 PMid:22493413

- Pettitt, A.R., Jackson, R., Carruthers,
S., Dodd, J., Dodd, S., Oates, M., Johnson, G.G., Schuh, A., Matutes,
E., Dearden, C.E., Catovsky, D., Radford, J.A., Bloor, A., Follows,
G.A., Devereux, S., Kruger, A., Blundell, J., Agrawal, S., Allsup, D.,
Proctor, S., Heartin, E., Oscier, D., Hamblin, T.J., Rawstron, A. and
Hillmen, P., Alemtuzumab in combination with methylprednisolone is a
highly effective induction regimen for patients with chronic
lymphocytic leukemia and deletion of TP53: final results of the
national cancer research institute CLL206 trial. J Clin Oncol 2012 30:
1647-1655. http://dx.doi.org/10.1200/JCO.2011.35.9695

- Stilgenbauer, S., Liebisch, P., James,
M.R., Schroder, M., Schlegelberger, B., Fischer, K., Bentz, M.,
Lichter, P. and Dohner, H., Molecular cytogenetic delineation of a
novel critical genomic region in chromosome bands 11q22.3-923.1 in
lymphoproliferative disorders. Proc Natl Acad Sci USA 1996 93:
11837-11841. http://dx.doi.org/10.1073/pnas.93.21.11837 PMid:10397742

- Schaffner, C., Stilgenbauer, S., Rappold,
G.A., Dohner, H. and Lichter, P., Somatic ATM mutations indicate a
pathogenic role of ATM in B-cell chronic lymphocytic leukemia. Blood
1999 94: 748-753. PMid:16014569

- Austen, B., Powell, J.E., Alvi, A.,
Edwards, I., Hooper, L., Starczynski, J., Taylor, A.M., Fegan, C.,
Moss, P. and Stankovic, T., Mutations in the ATM gene lead to impaired
overall and treatment-free survival that is independent of IGVH
mutation status in patients with B-CLL. Blood 2005 106: 3175-3182. http://dx.doi.org/10.1182/blood-2004-11-4516 PMid:21993670 PMCid:3248930

- Guarini, A., Marinelli, M., Tavolaro, S.,
Bellacchio, E., Magliozzi, M., Chiaretti, S., De Propris, M.S.,
Peragine, N., Santangelo, S., Paoloni, F., Nanni, M., Del Giudice, I.,
Mauro, F.R., Torrente, I. and Foa, R., ATM gene alterations in chronic
lymphocytic leukemia patients induce a distinct gene expression profile
and predict disease progression. Haematologica 2012 97: 47-55. http://dx.doi.org/10.3324/haematol.2011.049270 PMid:22952040

- Ouillette, P., Li, J., Shaknovich, R., Li,
Y., Melnick, A., Shedden, K. and Malek, S.N., Incidence and clinical
implications of ATM aberrations in chronic lymphocytic leukemia. Genes
Chromosomes Cancer 2012 http://dx.doi.org/10.1002/gcc.21997 PMid:21266718

- Rossi, D., Spina, V., Deambrogi, C., Rasi,
S., Laurenti, L., Stamatopoulos, K., Arcaini, L., Lucioni, M., Rocque,
G.B., Xu-Monette, Z.Y., Visco, C., Chang, J., Chigrinova, E., Forconi,
F., Marasca, R., Besson, C., Papadaki, T., Paulli, M., Larocca, L.M.,
Pileri, S.A., Gattei, V., Bertoni, F., Foa, R., Young, K.H. and
Gaidano, G., The genetics of Richter syndrome reveals disease
heterogeneity and predicts survival after transformation. Blood 2011
117: 3391-3401. http://dx.doi.org/10.1182/blood-2010-09-302174 PMid:22275381

- Xu-Monette, Z.Y., Medeiros, L.J., Li, Y.,
Orlowski, R.Z., Andreeff, M., Bueso-Ramos, C.E., Greiner, T.C.,
McDonnell, T.J. and Young, K.H., Dysfunction of the TP53 tumor
suppressor gene in lymphoid malignancies. Blood 2012 119: 3668-3683. http://dx.doi.org/10.1182/blood-2011-11-366062 PMid:20861914

- Zenz, T., Vollmer, D., Trbusek, M.,
Smardova, J., Benner, A., Soussi, T., Helfrich, H., Heuberger, M.,
Hoth, P., Fuge, M., Denzel, T., Habe, S., Malcikova, J., Kuglik, P.,
Truong, S., Patten, N., Wu, L., Oscier, D., Ibbotson, R., Gardiner, A.,
Tracy, I., Lin, K., Pettitt, A., Pospisilova, S., Mayer, J., Hallek,
M., Dohner, H., Stilgenbauer, S. and European Research Initiative on,
C.L.L., TP53 mutation profile in chronic lymphocytic leukemia: evidence
for a disease specific profile from a comprehensive analysis of 268
mutations. Leukemia 2010 24: 2072-2079. http://dx.doi.org/10.1038/leu.2010.208 PMid:22297721

- Pospisilova, S., Gonzalez, D., Malcikova,
J., Trbusek, M., Rossi, D., Kater, A.P., Cymbalista, F., Eichhorst, B.,
Hallek, M., Dohner, H., Hillmen, P., van Oers, M., Gribben, J., Ghia,
P., Montserrat, E., Stilgenbauer, S., Zenz, T. and European Research
Initiative on, C.L.L., ERIC recommendations on TP53 mutation analysis
in chronic lymphocytic leukemia. Leukemia 2012 26: 1458-1461. http://dx.doi.org/10.1038/leu.2012.25 PMid:19414856

- Tam, C.S., Shanafelt, T.D., Wierda, W.G.,
Abruzzo, L.V., Van Dyke, D.L., O'Brien, S., Ferrajoli, A., Lerner,
S.A., Lynn, A., Kay, N.E. and Keating, M.J., De novo deletion 17p13.1
chronic lymphocytic leukemia shows significant clinical heterogeneity:
the M. D. Anderson and Mayo Clinic experience. Blood 2009 114: 957-964.
http://dx.doi.org/10.1182/blood-2009-03-210591 PMid:20595516

- Dreger, P., Dohner, H., Ritgen, M.,
Bottcher, S., Busch, R., Dietrich, S., Bunjes, D., Cohen, S., Schubert,
J., Hegenbart, U., Beelen, D., Zeis, M., Stadler, M., Hasenkamp, J.,
Uharek, L., Scheid, C., Humpe, A., Zenz, T., Winkler, D., Hallek, M.,
Kneba, M., Schmitz, N., Stilgenbauer, S. and German, C.L.L.S.G.,
Allogeneic stem cell transplantation provides durable disease control
in poor-risk chronic lymphocytic leukemia: long-term clinical and MRD
results of the German CLL Study Group CLL3X trial. Blood 2010 116:
2438-2447. http://dx.doi.org/10.1182/blood-2010-03-275420 PMid:20967796 PMCid:2996483

- Aster, J.C., Blacklow, S.C. and Pear,
W.S., Notch signalling in T-cell lymphoblastic leukaemia/lymphoma and
other haematological malignancies. J Pathol 2011 223: 262-273. http://dx.doi.org/10.1002/path.2789 PMid:21948802 PMCid:3182047

- Lobry, C., Oh, P. and Aifantis, I.,
Oncogenic and tumor suppressor functions of Notch in cancer: it's NOTCH
what you think. J Exp Med 2011 208: 1931-1935. http://dx.doi.org/10.1084/jem.20111855 PMid:21646301 PMCid:3111953

- Rothenberg, E.V., T cell lineage commitment: identity and renunciation. J Immunol 2011 186: 6649-6655. http://dx.doi.org/10.4049/jimmunol.1003703 PMid:17878313 PMCid:2000509

- Santos, M.A., Sarmento, L.M., Rebelo, M.,
Doce, A.A., Maillard, I., Dumortier, A., Neves, H., Radtke, F., Pear,
W.S., Parreira, L. and Demengeot, J., Notch1 engagement by Delta-like-1
promotes differentiation of B lymphocytes to antibody-secreting cells.
Proc Natl Acad Sci USA 2007 104: 15454-15459. http://dx.doi.org/10.1073/pnas.0702891104 PMid:22086416

- Balatti, V., Bottoni, A., Palamarchuk, A.,
Alder, H., Rassenti, L.Z., Kipps, T.J., Pekarsky, Y. and Croce, C.M.,
NOTCH1 mutations in CLL associated with trisomy 12. Blood 2012 119:
329-331. http://dx.doi.org/10.1182/blood-2011-10-386144 PMid:22207691 PMCid:3291600

- Del Giudice, I., Rossi, D., Chiaretti, S.,
Marinelli, M., Tavolaro, S., Gabrielli, S., Laurenti, L., Marasca, R.,
Rasi, S., Fangazio, M., Guarini, A., Gaidano, G. and Foa, R., NOTCH1
mutations in +12 chronic lymphocytic leukemia (CLL) confer an
unfavorable prognosis, induce a distinctive transcriptional profiling
and refine the intermediate prognosis of +12 CLL. Haematologica 2012
97: 437-441. http://dx.doi.org/10.3324/haematol.2011.060129 PMid:22619094

- Lopez, C., Delgado, J., Costa, D., Conde,
L., Ghita, G., Villamor, N., Navarro, A., Cazorla, M., Gomez, C.,
Arias, A., Munoz, C., Baumann, T., Rozman, M., Aymerich, M., Colomer,
D., Cobo, F., Campo, E., Lopez-Guillermo, A., Montserrat, E. and
Carrio, A., Different distribution of NOTCH1 mutations in chronic
lymphocytic leukemia with isolated trisomy 12 or associated with other
chromosomal alterations. Genes Chromosomes Cancer 2012 51: 881-889. http://dx.doi.org/10.1002/gcc.21972 PMCid:3415400

- Groth, C. and Fortini, M.E., Therapeutic
approaches to modulating Notch signaling: current challenges and future
prospects. Semin Cell Dev Biol 2012 23: 465-472. http://dx.doi.org/10.1016/j.semcdb.2012.01.016 PMid:20965628 PMCid:3033461

- Paganin, M. and Ferrando, A., Molecular
pathogenesis and targeted therapies for NOTCH1-induced T-cell acute
lymphoblastic leukemia. Blood Rev 2011 25: 83-90. http://dx.doi.org/10.1016/j.blre.2010.09.004

- Will, C.L. and Luhrmann, R., Spliceosome structure and function. Cold Spring Harb Perspect Biol 2011 3: http://dx.doi.org/10.1101/cshperspect.a003707 PMid:23142775 PMCid:3493507

- David, C.J. and Manley, J.L., Alternative
pre-mRNA splicing regulation in cancer: pathways and programs unhinged.
Gene Dev 2010 24: 2343-2364. http://dx.doi.org/10.1101/gad.1973010 PMid:8649382 PMCid:231265

- Luke, M.M., Della Seta, F., Di Como, C.J.,
Sugimoto, H., Kobayashi, R. and Arndt, K.T., The SAP, a new family of
proteins, associate and function positively with the SIT4 phosphatase.
Mol Cell Biol 1996 16: 2744-2755.

- Wang, C., Chua, K., Seghezzi, W., Lees,
E., Gozani, O. and Reed, R., Phosphorylation of spliceosomal protein
SAP 155 coupled with splicing catalysis. Gene Dev 1998 12: 1409-1414.
PMid:10490618 PMCid:84676

- Das, B.K., Xia, L., Palandjian, L.,
Gozani, O., Chyung, Y. and Reed, R., Characterization of a protein
complex containing spliceosomal proteins SAPs 49, 130, 145, and 155.
Mol Cell Biol 1999 19: 6796-6802. PMid:19239890

- Wahl, M.C., Will, C.L. and Luhrmann, R., The spliceosome: design principles of a dynamic RNP machine. Cell 2009 136: 701-718. http://dx.doi.org/10.1016/j.cell.2009.02.009 PMid:21995386 PMCid:3322589

- Papaemmanuil, E., Cazzola, M., Boultwood,
J., Malcovati, L., Vyas, P., Bowen, D., Pellagatti, A., Wainscoat,
J.S., Hellstrom-Lindberg, E., Gambacorti-Passerini, C., Godfrey, A.L.,
Rapado, I., Cvejic, A., Rance, R., McGee, C., Ellis, P., Mudie, L.J.,
Stephens, P.J., McLaren, S., Massie, C.E., Tarpey, P.S., Varela, I.,
Nik-Zainal, S., Davies, H.R., Shlien, A., Jones, D., Raine, K., Hinton,
J., Butler, A.P., Teague, J.W., Baxter, E.J., Score, J., Galli, A.,
Della Porta, M.G., Travaglino, E., Groves, M., Tauro, S., Munshi, N.C.,
Anderson, K.C., El-Naggar, A., Fischer, A., Mustonen, V., Warren, A.J.,
Cross, N.C., Green, A.R., Futreal, P.A., Stratton, M.R., Campbell, P.J.
and Chronic Myeloid Disorders Working Group of the International Cancer
Genome, C., Somatic SF3B1 mutation in myelodysplasia with ring
sideroblasts. N Engl J Med 2011 365: 1384-1395. http://dx.doi.org/10.1056/NEJMoa1103283 PMid:11907583

- Li, X., Yang, Y. and Ashwell, J.D.,
TNF-RII and c-IAP1 mediate ubiquitination and degradation of TRAF2.
Nature 2002 416: 345-347. http://dx.doi.org/10.1038/416345a PMid:18997794 PMCid:2676931

- Zarnegar, B.J., Wang, Y., Mahoney, D.J.,
Dempsey, P.W., Cheung, H.H., He, J., Shiba, T., Yang, X., Yeh, W.C.,
Mak, T.W., Korneluk, R.G. and Cheng, G., Noncanonical NF-kappaB
activation requires coordinated assembly of a regulatory complex of the
adaptors cIAP1, cIAP2, TRAF2 and TRAF3 and the kinase NIK. Nature
Immunol 2008 9: 1371-1378. http://dx.doi.org/10.1038/ni.1676

- Vallabhapurapu, S. and Karin, M.,
Regulation and function of NF-kappaB transcription factors in the
immune system. Mol Cell Biol 2009 27: 693-733. http://dx.doi.org/10.1146/annurev.immunol.021908.132641 PMid:21048983 PMCid:2964333

- Conze, D.B., Zhao, Y. and Ashwell, J.D.,
Non-canonical NF-kappaB activation and abnormal B cell accumulation in
mice expressing ubiquitin protein ligase-inactive c-IAP2. PLoS Biol
2010 8: e1000518. http://dx.doi.org/10.1371/journal.pbio.1000518

- Hewamana, S., Lin, T.T., Jenkins, C.,
Burnett, A.K., Jordan, C.T., Fegan, C., Brennan, P., Rowntree, C. and
Pepper, C., The novel nuclear factor-kappaB inhibitor LC-1 is
equipotent in poor prognostic subsets of chronic lymphocytic leukemia
and shows strong synergy with fludarabine. Clinl Cancer Res 2008 14:
8102-8111. http://dx.doi.org/10.1158/1078-0432.CCR-08-1673 PMid:20351313 PMCid:2904580

- Hertlein, E., Wagner, A.J., Jones, J.,
Lin, T.S., Maddocks, K.J., Towns, W.H., 3rd, Goettl, V.M., Zhang, X.,
Jarjoura, D., Raymond, C.A., West, D.A., Croce, C.M., Byrd, J.C. and
Johnson, A.J., 17-DMAG targets the nuclear factor-kappaB family of
proteins to induce apoptosis in chronic lymphocytic leukemia: clinical
implications of HSP90 inhibition. Blood 2010 116: 45-53. http://dx.doi.org/10.1182/blood-2010-01-263756
