Received: November 11, 2013
Accepted: December 13, 2013
Meditter J Hematol Infect Dis 2014, 6(1): e2013004, DOI 10.4084/MJHID.2014.004
This article is available on PDF format at:

Tathagata Chatterjee1, Srishti Gupta2, Sanjeevan Sharma3 and Prosenjit Ganguli4
1 Professor
& Head of Department, Department of Immunohematology and
Transfusion Medicine, Armed Forces Medical College, Pune, India.
2 Senior Resident, Department of Pathology, NDMC
Medical College, Delhi, India.
3 Associate Professor,
Department of Haematology, Army Hospital(R & R, Delhi, India.
4 Associate Professor, Department of Pathology,
Army Hospital(R & R), Delhi, India.

| This is an Open Access article distributed
under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0),
which permits unrestricted use, distribution, and reproduction in any
medium, provided the original work is properly cited. |
|
Abstract Context: Acute promyelocytic leukemia (APL), an AML subtype, is characterized morphologically by abnormal promyelocytes. Molecular studies show three possible bcr isoforms of PML-RARα fusion gene. This study undertakes analysis of PML-RARα bcr isoforms and their correlation with haematological parameters and response to treatment in Indian patients.Aims: To study different PML-RARα bcr isoforms in Indian patients and to find any correlation with various haematological parameters and response to treatment Settings and Design: Patients diagnosed as APL on morphology or flowcytometry and confirmed by RQ PCR were included in the study. Treated APL patients or patients with relapse and on follow-up were excluded from the study. Methods and Material: Twenty patients over thirty one months period were included. The clinical, haematological & morphological features were analysed, the latter using routine & special cytochemical stains on blood and bone marrow. Flow cytometric evaluation using 4-color Beckman Coulter FC 500 and molecular studies using RT PCR Fusion Quant® kits for bcr-1, bcr-2 and bcr-3 of PML- RARα bcr isoforms on the instrument Rotor Gene™ 3000 were performed. Statistical analysis used: Student t test was applied to correlate different bcr isoforms with various haematological parameters and response to treatment. Results: In our study, M:F ratio was 1.5:1 with median age 42 years, Hb - 8.0 g/dl, TLC-7900/μl, and platelet – 35000/ μl and varied clinical presentation. Four patients were microgranular variants, and the rest were hypergranular. MPO and CAE positivity were100% and for NSE it was 33.33%. Molecular analysis revealed PML-RARα isoforms of bcr1 in 42.85%, bcr2 in 14.28% and bcr3 in 38.09% patients. No correlation was found between PML-RARα bcr isoforms, different haematological parameters and response to treatment. Conclusions: Higher incidence of PML- RARα bcr-1 isoform was found in Indian APL patients with no significant correlation between different haematological parameters and response to treatment. |
Introduction
Acute
promyelocytic leukemia is a distinct subtype of acute myeloid leukemia
(AML), which occurs in 5-13% of patients diagnosed with AML.[1-3]
Morphologically the bone marrow shows effacement by heavily granulated
cells with folded twisted nuclei. Cytogenetically, it is characterized
by balanced reciprocal translocation between chromosome 15 and 17 which
results in fusion between promyelocytic leukemia (PML) gene and
Retinoic acid receptor α (RARα) gene.[1]
There are 3 possible PML-RARα isoforms caused by these translocations.
The breakpoint in chromosome 17 is consistently found in intron 2, but
varies in chromosome 15. The 3 breakpoints on the PML gene can occur at
intron 3 (L-long form), intron 6 (S-short form), and exon 6 (V form).[4-6]
It has been reported that the S (short) form is associated with a
shorter remission duration and overall survival compared with the L
form in a study by Gonzales et al..[5]
This
translocation can be detected by karyotyping or fluorescence in situ
hybridization (FISH) studies, and the transcript can be detected by
molecular techniques like the polymerase chain reaction (PCR)
techniques. The distribution of the breakpoint sites in the PML gene
has been reported in several studies from Europe and USA to be
approximately 50–55% for PML (L) - RARα, 8–20% for PML (V)-RARα and
27–49% for PML (S)-RARα.[7] In a
study by Dutta et al from India bcr 3 was found to be predominant.[8]
Reports suggesting an association between different PML- RARα bcr
breakpoint sites and clinical characteristics or response to treatment
in APL patients have not been consistent.[5-7]
Subjects and methods
We analyzed twenty APL patients confirmed by RQ-PCR in the
study during
the period March 2010 to September 2012. All the patients fulfilled
morphologic criteria for APL (Hypergranular and microgranular variant)
as classified by WHO classification.[9]
In addition,
all patients manifested the PML/RARα rearrangements by RQ PCR. Patients
with presumptive morphologic diagnosis but negative molecular result
were excluded from this study. Clinical features, hematological
parameters [Hemoglobin (Hb), total leukocyte count (TLC), platelet
count, prothrombin time and activated partial thromboplastin time,
D-dimer and fibrinogen] were analyzed at presentation.
Patients
were grouped under low risk group (TLC <10,000/ μl,
platelet
>40,000/ μl), intermediate risk group (TLC <10,000/ μl,
platelet
count < 40,000/ μl) and high risk group (TLC >10,000/
μl).[10]
Peripheral blood and bone marrow aspirate were stained with Leishman
stain and cytochemistry consisted of myeloperoxidase (MPO),
Chloroacetate esterase (CAE) and non-specific esterase (NSE) using
Merck’s® Diagnostic reagents and manufacturer specified staining
instructions.
Flow cytometric evaluation was done on bone marrow aspirate using
Beckman Coulter FC 500 four colour flowcytometer using standard Lyse
wash technique. It was done on eleven patients only due to
nonavailability of reagents. Antibodies used were labelled with
FITC,PE,ECD and PC-5. Antibody panel used was CD13, CD33, CD45, cMPO,
cCD79a, cCD3, CD15, HLA-DR, CD10, CD19, CD34, CD7, CD117 and sCD3.
Antibody was considered positive if more than 20% of cells gated were
positive. Sample was processed within 24 hours of collection. Gating
strategy used was CD45 vs. SSC.
Samples were outsourced for cytogenetic evaluation due to lack of
facilities in the centre for balanced reciprocal translocation between
chromosome 15 and chromosome 17.
For molecular studies, peripheral blood sample was collected. RQ PCR
was done for PML-RARα fusion transcript. RNA was extracted and
converted to cDNA using Applied Biosystems® High Capacity cDNA Reverse
transcription kit on the same day of RNA extraction. cDNA was amplified
using Fusion Quant® kits for PML (L)-RARα, PML (V)-RARα and PML(S)-RARα
on the instrument Rotor Gene™ 3000 as per the manufacturer’s
instructions. ABL transcript was used as an endogenous control gene.
Water was used as a negative control. The test samples were run in
duplicate.
RQ – PCR was carried out using Taqman® universal PCR Master Mix and
IPSOGEN® Primers and probe mix. Three different probes and primers were
run for PML (L) - RARα, PML (V)-RARα and PML(S)-RARα all of which had
separate kits. The kit used for PMLRARAα is a quantitative individual
primer based one and not multiplexes. Unfortunately, the only kit
available as a multiplex one in our country is a qualitative one. The
kit used in our dept is tabulated below.
 |
The standards of
each are labelled as F1, F2, F3, F4, F5. These have 10,100,1000,100,000
and 10,00,000 copies/5ul respectively.
Initially, we ran control gene, in our case ABL gene which has three
standards C1, C2, C3 of 1000, 10,000 and 100,000 copies/5ul
respectively. If these were in acceptable range then we proceeded for
PML(L)- RARα, PML(V)-RARα and PML(S)-RARα separately along
with
their 5 standards in a single Real time Q PCR and not multiplex.
Total number of cycles run was 45 cycles over a period of 2 hours and
20 minutes. Standard curves were obtained for control gene, fusion gene
of standard and test samples after acquisition
The Control Gene (CG) standard curve equation was used to transform raw
cycle threshold (Ct) values obtained with Primer & Probe Mix
for
Control gene (CG- PPC) for the unknown samples, into the CG copy
numbers (CG CN).
The Fusion Gene standard curve equation was used to transform raw Ct
values (obtained with Fusion gene Primer & Probe Mix (FG- PPF)
for
the unknown samples into Fusion Gene copy numbers (FG CN). The ratio of
these copy numbers gives the normalized copy number (NCN):
NCN = FG CN / CG CN
Any value of normalized copy number above zero was considered as
significant.
Patients were treated with all Trans retinoic acid (ATRA) based
regimen. Consolidation strategies adopted in our Institute depended on
the risk classification for relapse at diagnosis. Two or 3 cycles of
anthracycline-based chemotherapy were administered for low- and
intermediate-risk patients in first complete remission (CR1) as shown
in table 1.
For patients with
high-risk disease (WBC>10 000/μL), either intermediate- or
high-dose
Ara-C was administered depending on age as a first consolidation as
done in the GIMEMA APL 2000 study or ATO for 2 courses of 25 days each
as carried out in the second North American Intergroup C9710 study.
Statistical analysis: Results were statistically
analyzed using
SPSS software (version 17.0, SPSS, Chicago, Illinois, USA). Concordance
rates were calculated between aspirate and biopsy. In groups with
sufficient sample size, validity parameters were calculated. Proportion
of trephine biopsies showing lymphoma infiltration was plotted on y
axis and total preprocessing trephine length on x axis, in increments
of 4 mm. Fischer’s exact test was used to analyze the significance in
lymphoma positivity between two groups and P value <0.05 was
considered statistically significant.
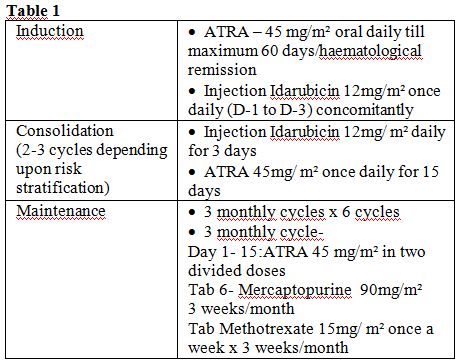 |
Table 1 |
Results
Twenty patients were studied. Patient characteristics are
given in table 2.
The
presenting clinical features are given in table 3.
On morphological analysis, four patients were microgranular variants
and sixteen were hypergranular. MPO and CAE stains were seen positive
in 100% of APL cases, and NSE was seen in 33.33% of cases.
Flowcytometric analysis of eleven patients [microgranular- n (2),
hypergranular-n (9)] revealed characteristic findings of APL.[11]
All hypergranular variants revealed a high side scatter and antigenic
expression of CD13, CD33 and MPO. CD117 was expressed in one
hypogranular and two hypergranular variants. Weak CD34 expression was
seen in two hypogranular variants. HLA-DR expression was not seen in
any case. Weak CD15 was noted in three hypergranular variants.
All the patients were cytogenetically determined to be characteristic t
(15; 17) (q22; q21).
Molecular analysis revealed PML(L) RARα in 42.85%, PML(V) RARα in
14.28% and PML(S) RARα in 38.09% patients. Amongst these, two
microgranular variants were PML(L) RARα positive, third microgranular
variant was PML(V) RARα and the fourth PML(S) RARα positive. Five
patients died during the induction phase of which three were PML(S)
RARα positive and two were PML(L) RARα positive. Three patients died
due to intracranial haemorrhage, and two patients died due to
hyperkalemia leading to cardiac arrest. Rest of the patients achieved
molecular remission. No significant correlation was found between
PML-RARα bcr isoforms, age, sex and different haematological parameters
namely haemoglobin, total leukocyte count, platelet count and response
to treatment.
| Table 2. |
| Table 3. |
Discussion
On examination, one unusual feature was gum hypertrophy seen
in four
patients (20%). This observation is similar to a previous Indian study
by Dutta et al.[8] where the same
clinical feature was
observed in some of their patients. However unlike that study not a
single case of ours presented with scrotal ulceration. We feel that
presence of gum hypertrophy seen commonly in acute myelomonocytic
leukemia is also seen in Indian APL patients. Generally, gum
hypertrophy is very rarely seen in AML-M3.[12]
Another unusual feature was high TLC at presentation. TLC varied from
3000/μl to 71,000/μl with a median of 7900/μl. APL is characterised by
pancytopenia in peripheral blood. Raised TLC is seen in 10-30% of cases
especially with microgranular variant.[12]
In our
series of twenty patients, four were microgranular variants (20%).
Three out of four patients of microgranular variants showed high TLC at
presentation. Statistical significance was not determined due to the
small number of cases.
Cytochemistry results showed positivity for MPO and CAE in all the APL
cases (100%). Interestingly 33.33% of cases also showed positivity for
NSE. This finding is in concordance with previous published literature
where NSE positivity in APL cases is between 13.5% - 60.7%.[8,13]
RQ-PCR done at the time of diagnosis revealed PML-RARα transcript in
twenty patients. Unusual feature was that in our study PML(L)
RARα isoform was found to be the predominant isoform (42.85%)
followed by PML(S) RARα isoform (38.09%). Whereas according to data
published in India by Dutta et al.[8]
PML(S) RARα
isoform was found to be significantly high (72.7%). However, our study
results were in concordance with published western studies.[7] In a study of 2003 Douer et al.[7]
found a frequency of PML(L) RARα isoform significantly high.
They
summarized also the distribution of the breakpoint sites in the PML
gene reported in several studies from Europe and the USA, which was
approximately 50–55% for PML(L) RARα, 8–20% for PML(V) RARα and
27–49% for PML(S) RARα.[7]
This finding suggests
that PML (L) RARα subtype may represent a distinct biological subset
and breakpoint at intron 6 in PML gene may not be a random event. This
might be possibly related to genetic and/ or environmental factor(s)
playing a role in determining the breakage site of the PML gene.
Multicentre trial studies are needed to confirm the same.
Of total twenty patients in the study, five patients (25%) died during
the induction phase. Three were PML(S) RARα positive and two were
PML(L) RARα positive. No mortality was noted with PML(V) RARα
isoform. In previous studies of cytotoxic chemotherapy, early deaths
during induction in patients with APL occurred primarily as a
consequence of intracranial hemorrhage.[14,15]
The
hemorrhagic diathesis of APL is related to depletion of platelets and
clotting factors, probably owing to leukemic cell lysis and release of
procoagulant or fibrinolytic materials into the circulation.[16,17] Early mortality from this
problem has ranged from 10% to 47% in published series.[18,19]
According to published studies PML(S) RARα isoform has been related to
inferior duration of remission and overall survival.[20]
All our 15 patients are under regular follow-up. There is a system in
the Indian Armed Forces patients to compulsorily attend follow-up as
per standard instructions of the Clinicians. A separate long-term
follow-up study is being devised by our Clinical
investigators
for all APL patients and is likely to be published in two years time
involving at least 50 patients; hence it will be beyond the scope of
this particular publication to consider the over-all DFS, median FU and
OS. However, regular follow-up is being meticulously recorded for all
our fifteen APL patients.
No significant correlation was found between the PML-RARα isoforms,
haematological parameters, age, sex and response to treatment; however
study involving a small sample size may not have the power to detect
such a relationship. This is similar to results of previous studies.[7,21]
However some studies have found an association between PML(S)
RARα isoform and high TLC[22]
and PML(V) RARα isoform and high TLC at presentation.[5]
Conclusion
To conclude, APL is a distinct biological entity. In our study PML(L) RARα isoform was found to be the predominant isoform (in concordance with western studies) followed by PML(S) RARα isoform with no significant correlation with age, sex, haematological parameters and response to treatment.
References



















[TOP]