Diagnosis and Subclassification of Acute Lymphoblastic
Leukemia
Sabina Chiaretti1,
Gina Zini2 and Renato Bassan3
1 Division
of Hematology, Department of Cellular Biotechnologies and Hematology,
“Sapienza” University of Rome, Rome, Italy
2 Hematology, Catholic University Sacred Heart
Policlinico Gemelli, Rome, Italy
3 Hematology and Bone Marrow Transplant Unit,
Ospedale dell’Angelo e SS. Giovanni e Paolo, Mestre-Venezia, Italy
Corresponding author: Sabina Chiaretti, Division of
Hematology, Department of Cellular Biotechnologies and
Hematology,“Sapienza” University of Rome, Via Benevento 6, 00161, Rome,
Italy. E-mail:
chiaretti@bce.uniroma1.it
Published: October 24, 2014
Received: September 18, 2014
Accepted: October 20, 2014
Meditter J Hematol Infect Dis 2014, 6(1): e2014073, DOI
10.4084/MJHID.2014.073
This article is available on PDF format at:

This is an Open Access article distributed
under the terms of the Creative Commons Attribution License
(http://creativecommons.org/licenses/by/2.0),
which permits unrestricted use, distribution, and reproduction in any
medium, provided the original work is properly cited.
|
|
Abstract
Acute lymphoblastic leukemia (ALL) is
a disseminated malignancy of B- or T-lymphoblasts which imposes a rapid
and accurate diagnostic process to support an optimal risk-oriented
therapy and thus increase the curability rate. The need for a precise
diagnostic algorithm is underlined by the awareness that both ALL
therapy and related success rates may vary greatly between ALL subsets,
from standard chemotherapy in patients with standard-risk ALL, to
allotransplantation (SCT) and targeted therapy in high-risk patients
and cases expressing suitable biological targets, respectively. This
review summarizes how best to identify ALL and the most relevant ALL
subsets. |
Introduction
Current standards for acute lymphoblastic leukemia (ALL) diagnosis
integrate the study of cell morphology, immunophenotype and
genetics/cytogenetics as detailed in the 2008 WHO classification of
lymphoid neoplasms.[1] The classification originally suggested by the FAB group is no longer followed.[2,3]
The FAB classification was clinically useful since it permitted
recognition of probable Burkitt lymphoma in leukemic phase, but it has
now been replaced by the WHO classification. Lymphoid neoplasms are
assigned, in the most recent WHO classification, to two principal
categories: neoplasms derived from B- and T-lineage lymphoid precursors
and those derived from mature B, T or NK cells. ALL belongs to the
first of these major groups, designated B- or T-lymphoblastic
leukemia/lymphoma[4] and including three principal
categories: B-lymphoblastic leukemia/lymphoma not otherwise specified,
B-lymphoblastic leukemia/lymphoma with recurrent cytogenetic
alterations and T-lymphoblastic leukemia/lymphoma. The designation of
leukemia/lymphoma reflects the principle that these neoplasms should be
classified on the basis of their biological and molecular
characteristics, regardless of the sites of involvement. The leukemic
variant shows diffuse involvement of the peripheral blood and the bone
marrow, while lymphoma is confined to nodal or extranodal sites, with
no or minimal involvement of the bone marrow. In the leukemic form, by
definition, the bone marrow must contain at least 20% blast cells. A
purely leukemic presentation is most typical of B-lineage ALL (85%),
while cases of T-lineage disease often present with an associated
lymphomatous mass in the mediastinum or other sites.
Diagnostic Morphology and Cytochemistry
A morphological bone marrow assessment represents the first step in
the diagnostic pathway, for the primary diagnosis of ALL and for the
differentiation from acute myeloid leukemia (AML),[5] since ALL, by definition, always presents with bone marrow involvement. Table 1[6]
shows the morphological criteria that are useful for distinguishing
between myeloblasts and lymphoblasts, however remembering the limits of
morphology in ALL, for which flow cytometry analysis represents the
diagnostic gold standard for both the identification of cell lineage
and the definition of subset. The morphology of leukemic cells in the
peripheral blood can be significantly different from that of the bone
marrow, which is always indispensable.
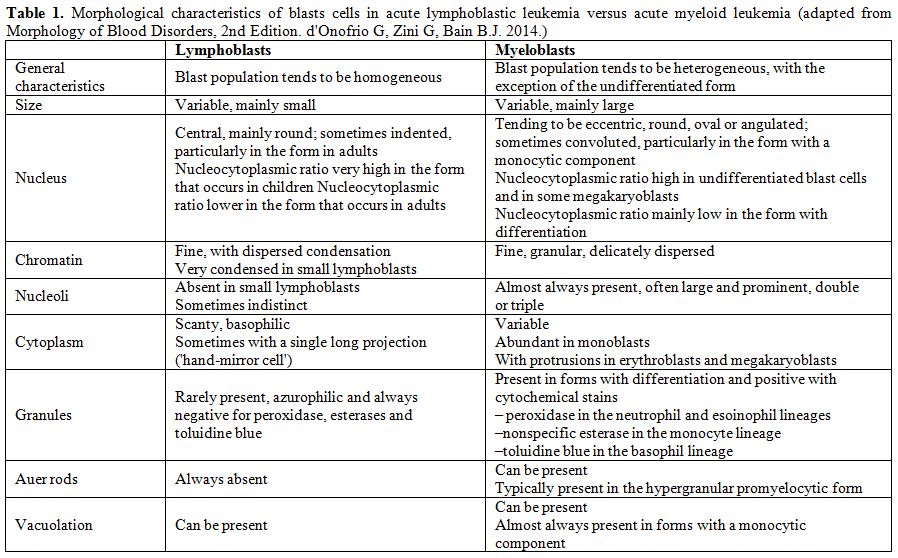 |
Table 1. Morphological characteristics of
blasts cells in acute lymphoblastic leukemia versus acute myeloid
leukemia (adapted from Morphology of Blood Disorders, 2nd Edition. d'Onofrio G, Zini G, Bain B.J. 2014). |
From the morphological point of view, there are no
reproducible criteria to distinguish between B- and T-lineage ALL. It
can also be difficult to distinguish B-lineage lymphoblasts from normal
B-lineage lymphoid precursors, known as hematogones, which are observed
in the peripheral blood in various conditions, including primary
myelofibrosis and in children in the phase of recovery following
chemotherapy.[7] Hematogones typically have an even
higher nucleocytoplasmic ratio than lymphoblasts, with more homogeneous
chromatin and a complete absence of visible nucleoli. Hematogones can
also express the CD10 antigen, but can be distinguished from blast
cells of B ALL by other immunophenotypic features, being characterised
by regular, orderly acquisition and loss of B-lineage antigens; they
can also be distinguished from mature lymphocytes by their weak
expression of CD45 and, sometimes, by the expression of CD34.[7]
The bone marrow morphology of ALL is however quite variable as previously indicated in the FAB classification (Figures 1-2).
Rare morphological variants are: ALL with “hand-mirror cells”, i.e. the
shape of the cells resembles a hand mirror or a tennis racquet (Figure 2A);
granular ALL, with presence of azurophilic cytoplasmic granules which
vary in number, size and shape. Cytochemically, these blasts have
negative peroxidase reactions and variable periodic acid-Schiff (PAS)
positivity; Sudan black B is sometimes weakly positive;[8]
ALL with mature cells that are nearly indistinguishable from mature
lymphoid neoplasms and require expert observers for accurate
morphological identification;[9] ALL associated with hypereosinophilia (Figure 2B). By definition, ALL blasts are negative for myeloperoxidase (MPO) (Figure 2C)
and other myeloid cytochemical reactions. According to the FAB
criteria, acute (leukemias with at least 3% MPO-positive blasts in BM
should be classified as myeloid. However, low level MPO positivity
without expression of other myeloid markers is detectable by means of
electron microscopy in rare ALL cases. True MPO+ ALL is discussed below
in the mixed lineage acute leukemias section. The acid phosphatase
reaction correlates with the lysosome content; it is useful for
identifying T-ALL blasts which show focal paranuclear positivity in
more than 80% of cases. Lymphoblasts may react with non-specific
esterases with a strong positivity in the Golgi zone with variable
inhibition with sodium fluoride. The B lymphoblasts in FAB L3/Burkitt
ALL show an intense cytoplasmic positivity to methyl green pyronine,
while the vacuoles stain strongly with Oil red O, thus demonstrating
their lipid content. The role of cytochemistry in differentiating ALL
from AML is limited and is mainly of historical interest, since these
tests have now been superseded by the far more objective results
provided by the immunophenotyping.
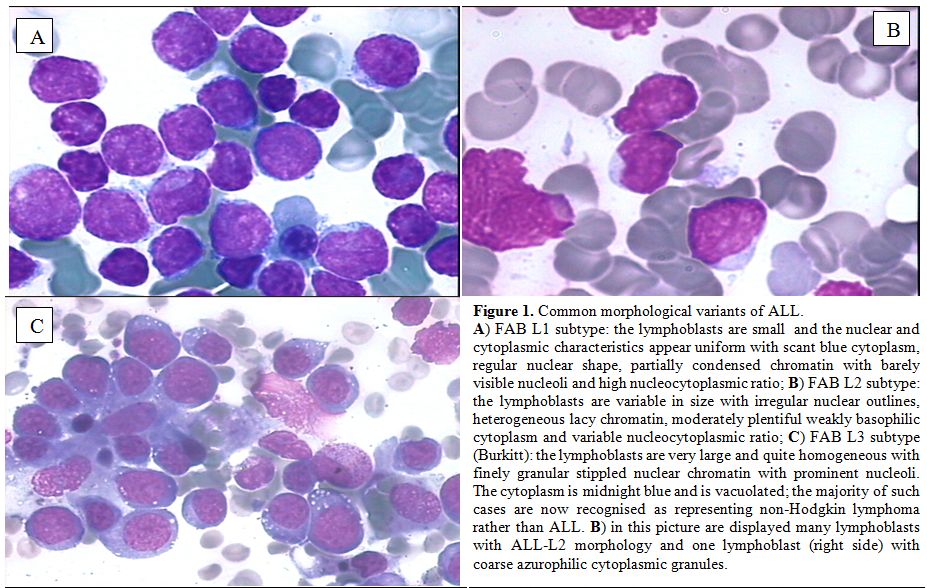 |
Figure 1. Common morphological variants of ALL.
A)
FAB L1 subtype: the lymphoblasts are small and the nuclear and
cytoplasmic characteristics appear uniform with scant blue cytoplasm,
regular nuclear shape, partially condensed chromatin with barely
visible nucleoli and high nucleocytoplasmic ratio; B) FAB L2 subtype:
the lymphoblasts are variable in size with irregular nuclear outlines,
heterogeneous lacy chromatin, moderately plentiful weakly basophilic
cytoplasm and variable nucleocytoplasmic ratio; C) FAB L3 subtype
(Burkitt): the lymphoblasts are very large and quite homogeneous with
finely granular stippled nuclear chromatin with prominent nucleoli. The
cytoplasm is midnight blue and is vacuolated; the majority of such
cases are now recognised as representing non-Hodgkin lymphoma rather
than ALL. B) in this picture are displayed many lymphoblasts with
ALL-L2 morphology and one lymphoblast (right side) with coarse
azurophilic cytoplasmic granules. |
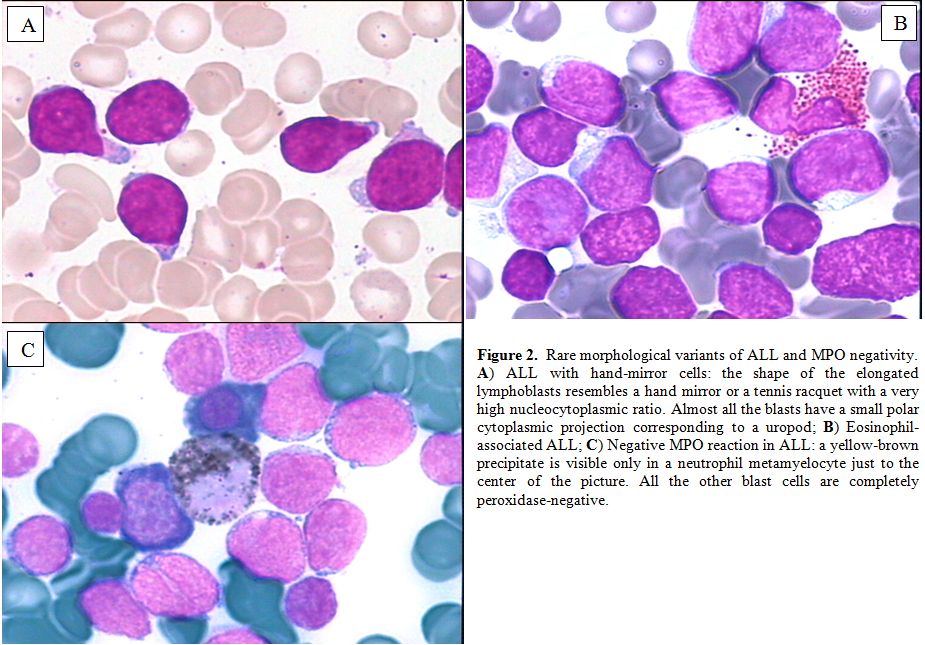 |
Figure 2. Rare morphological variants of ALL and MPO negativity.
A)
ALL with hand-mirror cells: the shape of the elongated lymphoblasts
resembles a hand mirror or a tennis racquet with a very high
nucleocytoplasmic ratio. Almost all the blasts have a small polar
cytoplasmic projection corresponding to a uropod;
B)
Eosinophil-associated ALL; C) Negative MPO reaction in ALL: a
yellow-brown precipitate is visible only in a neutrophil metamyelocyte
just to the center of the picture. All the other blast cells are
completely peroxidase-negative.
|
Diagnostic Morphology and Cytochemistry
Immunophenotyping by means of multi-channel flow cytometry (MFC) has
become the standard procedure for ALL diagnosis and subclassification,
and was also developed as useful tool for the detection and monitoring
of minimal residual disease (MRD, reviewed elsewhere in this issue).
The consensus by European Group for the Immunological characterization
of leukaemias (EGIL) is that a threshold of 20% should be used to
define a positive reaction of blast cells to a given monoclonal
antibody, except for MPO, CD3, CD79a and TdT, which are considered
positive at the 10% level of expression.[10,11] More
recently, novel MFC strategies were developed by the EuroFlow
consortium to ensure accurate methodologies through all MFC steps, in
order to guarantee the reproducibility of diagnostic tests.[12,13]
To summarize the diagnostic issue, roughly 75-80% of cases of adult ALL
are of B-cell lineage and 20-25% belong to the T-cell lineage.
Immunophenotype of B-lineage ALL
In
B-lineage ALL the most important markers for diagnosis, differential
diagnosis and subclassification are CD19, CD20, CD22, CD24, and CD79a.
The earliest B-lineage markers are CD19, CD22 (membrane and cytoplasm)
and CD79a.[14,15] A positive reaction for any two of
these three markers, without further differentiation markers,
identifies pro-B ALL (EGIL B-I subtype) (Figure 3A).
The presence of CD10 antigen (CALLA) defines the "common" ALL subgroup
(EGIL B-II subtype). Cases with additional identification of
cytoplasmic heavy mu chain constitute the pre-B group (EGIL B-III
subtype) (Figure 3B), whereas the presence of surface immunoglobulin light chains defines mature B-ALL (EGIL B-IV subtype). Among
other B-cell markers, B-I and B-II ALL are often CD24 positive and 4G7
(pro- and pre-B surrogate light chain specific MoAb) positive;[16]
surface CD20 and CD22 are variably positive beyond stage B-I; CD13 and
CD33 myeloid/cross lineage antigen can be expressed, as well as the
CD34 stem cell antigen, particularly in Ph+ (Philadelphia
chromosome-positive) ALL (often B-II with CD34, CD38, CD25 and
CD13/33), but myeloid-specific CD117 should not be present and can be
used to differentiate further between ALL and rare myeloid leukaemias
with negative MPO expression. Pro-B ALL with t(4;11)/MLL rearrangements
is most often myeloid antigen-positive disease (including expression of
CD15). TdT expression is usually lost in B-IV subgroup. T-cell markers
are usually not expressed in B-lineage ALL but a CD19+ subset is
concurrently CD2+. Loss of surface adhesion molecules has been
described.[17]
|
|
Figure
3. Examples of ALL immunophenotype.
A) Pro-B ALL: lymphoblasts are CD19, CD34, CD22, TdT and Cy CD79a positive and CD10 negative;
B)
Pre-B ALL: lymphoblasts are CD22, CD34, CD19, TdT, cytoplasmic
(Cy)CD79a, CD10 and Cy mμ positive; C) Cortical/thymic T-ALL:
Lymphoblasts are cyCD3, CD7, TdT, CD5, and CD1a positive.
|
Immunophenotype of T-lineage ALL
T-cell ALL
constitutes approximately 25% of all adult cases of ALL. T-cell markers
are CD1a, CD2, CD3 (membrane and cytoplasm), CD4, CD5, CD7 and CD8.
CD2, CD5 and CD7 antigens are markers of the most immature T-cell
cells, but none of them is absolutely lineage-specific, so that the
unequivocal diagnosis of T-ALL rests on the demonstration of
surface/cytoplasmic CD3. In T-ALL the expression of CD10 is quite
common (25%) and not specific; CD34 and myeloid antigens CD13 and/or
CD33 can be expressed too. Recognized T-ALL subsets are the following:
pro-T EGIL T-I (cCD3+, CD7+), pre-T EGIL T-II (cCD3+, CD7+ and
CD5/CD2+), cortical T EGIL T-III (cCD3+, Cd1a+, sCD3+/-) and mature-T
EGIL T-IV (cCD3+, sCD3+, CD1a-). Finally, a novel subgroup that was
recently characterized is represented by the so called ETP-ALL (Early-T
Precursor), which shows characteristic immunophenotypic features,
namely lack of CD1a and CD8 expression, weak CD5 expression, and
expression of at least one myeloid and/or stem cell marker.[18]
Mixed Phenotype Acute Leukemia
With
currently refined diagnostic techniques the occurrence of acute
leukemia of ambiguous cell lineage, i.e. mixed phenotype acute leukemia
(MPAL) is relatively rare (<4%).[19] These cases
express one of the following feature: 1) coexistence of two separate
blast cell populations (i.e. T- or B-cell ALL plus either myeloid or
monocytic blast cells, 2) single leukemic population of blast cells
co-expressing B- or T-cell antigens and myeloid antigens, 3) same plus
expression of monocytic antigens. For myelo-monocytic lineage useful
diagnostic antigens are MPO or nonspecific esterase, CD11c, CD14, CD64
and lysozyme; for B-lineage CD19 plus CD79a, cytoplasmic CD22 and CD10
(one or two of the latter according to staining intensity of CD19) and
for T-lineage cytoplasmic or surface CD3. Recognized entities include
Ph+ MPAL (B/myeloid or rarely T/myeloid), t(v;11q23;MLL rearranged
MPAL, and genetically uncharacterized B or T/myeloid MPAL. Very rare
cases express trilineage involvement (B/T/myeloid). Lack of
lineage-specific antigens (MPO, cCD3, cCD22) is observed in the
ultra-rare acute undifferentiated leukemia. In a recent review of 100
such cases,[20] 59% were B/myeloid, 35% T/myeloid, 4%
B/T lymphoid and 2% B/T/myeloid. Outcome was overall better following
ALL rather than AML therapy.
NK Cell ALL
CD56, a marker of natural killer
(NK) cell differentiation, defines a rare subgroup of about 3% of adult
ALL cases which often display other early T-cell antigens, CD7 CD2 CD5,
and sometimes cCD3.[19] True NK ALL is very rare (TdT+, CD56+, other T
markers negative, unrearranged TCR genes).[21] This diagnosis rely on the demonstration of early NK-specific CD94 or CD161 antigens.
Differential Diagnosis
With
few exceptions, ALL is readily identified by morphological marrow
assessment and MFC evaluation, with no need for additional tests, since
genetics/cytogenetics and genomics are available at a later stage and
cannot be employed for purely diagnostic purposes, even if they add
very useful clinical-prognostic information. Differentiation between
ALL and AML is initially obtained by excluding reactivity to SBB or MPO
stains in ALL cells (<3% positive). On cytochemical evaluation, some
rare ALL cases are SBB positive but MPO and chloroacetate esterase are
negative. True ALL cases that are immunoreactive to MPO or express
detectable levels of MPO mRNA have been described. This can occur in
Ph+ ALL and occasionally in T-lineage ALL.[22] Evaluation of CD117 antigen expression should also be carried out.[23]
Most ALL cases express the nuclear enzyme Terminal deoxynucleotidyl
Transferase (TdT). TdT-negative ALL is uncommonly reported, more in
T-ALL, while it is a rule in L3/Burkitt leukemia. Therefore all
TdT-negative B-precursor ALL cases must be thoroughly investigated to
exclude other aggressive lymphoid neoplasms with leukemic presentation
(blastic mantle cell lymphoma, atypical plasmablastic myeloma, other
high-grade lymphomas).[24,25]
Diagnostic Cytogenetics
Cytogenetics
represents an important step in ALL classification. Conventional
karyotyping can be helpful in the identification of recurrent
translocations, as well as gain and loss of gross chromosomal material;
however, the major limitation of this technique is that in some cases
leukemic cells fail to enter metaphase. However, fluorescence in situ
hybridization (FISH) can enable the detection and direct visualization
of virtually all investigated chromosomal abnormalities in ALL, with a
sensitivity of around 99%. Finally, array-comparative genomic
hybridization (array-CGH, a-CGH) and single nucleotide polymorphisms
(SNP) arrays can permit the identification of cryptic and/or
submicroscopic changes in the genome. Karyotypic changes found in ALL
include both numerical and structural alterations which have profound
prognostic significance.[26-30] With these premises
in mind, the karyotypic changes that occur in ALL can be roughly
subdivided in those associated respectively with a relatively good,
intermediate and poor prognosis (Table 2).[31-34]
However, it must be kept in mind that the incidence of certain
aberrations is very low, and that for some of them, the prognostic
impact can be strongly affected by the type and intensiveness of
therapy administered.
|
|
Table 2. Cytogenetics and prognosis in Ph-negative ALL.
Two karyotype-related prognostic classifications of Ph-negative ALL, as derived from two recent clinical series [31,32].
Definition of risk groups is according to the SWOG study, ranging from
<30% for the very high risk group to 50% and greater for the
favorable subtypes. Some differences are observed in the normal and
“other” karyotypic subgroups, which are assigned to the next better
category in the SWOG study compared to MRC-ECOG. It is necessary to
note that 9p deletions are not always associated with a favorable
prognosis. In a study identifying 18 such cases, survival was short and
comparable to Ph+ ALL [33].
|
Cytogenetic/Genetic Risk Groups
Among
the good prognosis aberrations, it is worth mentioning del(12p) or
t(12p)/t(12;21)(p13;q22) in B-lineage ALL, and t(10;14)(q24;q11) in
T-ALL. These abnormalities are relatively rare in adults compared with
childhood ALL.
Aberrations associated with an intermediate-risk
comprise the normal diploid subset plus cases with hyperdiploidy and
several other recurrent or random chromosomal abnormalities.
Other
aberrations, i.e. those with isolated trisomy 21, trisomy 8, and
perhaps del(6q) and t(1;19)(q23;p13)/E2A-PBX1 may constitute an
intermediate-high risk group; recent evidence suggests that the dismal
outcome previously reported for the t(1;19)(q23;p13)/E2A-PBX1 is
overcome by current therapeutic approaches.[35,36] Other recently identified aberrations in the intermediate high-risk group are represented by iAMP21[37] and IGH rearrangements, including CRLF2.[38]
Finally,
patients with t(9;22)(q34;q11)or BCR-ABL1 rearrangements or a positive
FISH test (Ph+ ALL), t(4;11)(q21;q23) or MLL rearrangements at 11q23,
monosomy 7, hypodiploidy/low hypodiploidy (and the strictly related
near triploid group) fall into the poor-risk cytogenetic category, with
an overall disease-free survival (DFS) rate of about 25%, or 10% in the
case of Ph+ ALL prior to the introduction of tyrosine kinase inhibitors
(TKI).[39-42] Ph+ ALL may constitute 25-50% of CD10+
common or pre-B ALL cases and represent the most frequent abnormality
in the adult/elderly, being detected in more than 50% of cases in 6th decade of life.[43] Secondary chromosome abnormalities in addition to t(9;22)(q34;q11) may worsen the prognosis;[44] however, this is as yet unproven in TKI era.[45]
Currently, the most unfavorable group within cases with known
genetic/molecular aberration is represented by t(4;11)(q21;q23) +
MLL1-rearranged ALL, for which outcome is very poor unless allogeneic
transplantation is adopted.[46]
Some other
karyotypes are unique to specific ALL syndromes. Translocations
involving chromosome 8 (MYC gene), such as t(8;14)(q24;q32) (90% of
cases), t(8;22)(q24;q11)(10% of cases), and t(2;8) (rarely observed),
are virtually present in 100% of cases of mature B-ALL with L3/Burkitt
morphology and clonal surface immunoglobulins. Typical cytogenetic
aberrations are also found in T-lineage ALL.[47] The
most frequent involve 14q11 breakpoints e.g. t(10;14)(q24;q11),
t(11;14)(p13;q11), or other. The presence of t(8;14) with breakpoints
at q24;q11 (q24;q32 in B-ALL) in T-ALL is associated with a
lymphomatous, aggressive presentation.[48,49]
New Genetics and Genomics in ALL
The
integration of results of several techniques, i.e. gene expression
profiling (GEP), SNP array analysis, and currently next-generation
sequencing (NGS), have permitted a better definition of the molecular
scenario of ALL and the identification of a constellation of novel
mutations; as for the latter, however, caution must be shown, since
while the biological role has been elucidated for some, while further
investigation is required for others. These findings are detailed below
(Tables 3, 4).
|
|
Table 3. Identification of novel lesions by integrated molecular genetics. |
|
|
Table 4. Summary of recurrent genetic lesions and mutations in T-ALL. |
B-lineage ALL: IKZF1, encoding for the transcription factor Ikaros, is frequently disrupted in BCR/ABL+ ALL (80% of cases). IKZF1 deletions, that can be different in size, are predictors of poor outcome in Ph+ ALL,[50-52] as well as in non-Ph+ ALL.[53-55]
Deregulated overexpression of CRLF2 (CRLF2), found exclusively in 5-10% B-ALL cases without known molecular rearrangements[56,57] is usually sustained by two types of aberrations: a rearrangement that involves CRLF2 and the Ig heavy chain locus (IGH@-CRLF2) or an interstitial PAR1 deletion that juxtaposes intron 1 of P2RY8 to the coding region of CRLF2 itself. More rarely, CRLF2 mutations can be detected. -CRLF2 can be detected together with IKZF1 deletion in Ph-negative ALL patients and with JAK mutations (JAK1 or JAK2) or IL7R mutations; furthermore, they are identified in roughly 50% of children with Down syndrome;[55,58] although some contrasting results have been reported, its presence correlates with an overall poor outcome.[54,55]
By the integration of genome-wide technologies, the “BCR/ABL-like” subgroup has been suggested/identified in both the adult[59,60] and pediatric populations[61,62]
and it accounts for about 15% of B-ALL cases. This subgroup is
characterized by a gene expression signature that is similar to that of
BCR/ABL+ patients, frequent detection of IKZF1 deletions and CRLF2
rearrangements and adismal outcome. NGS has revealed the presence of
mutations and/or rearrangements activating tyrosine kinases, i.e IGH-CRLF2, NUP214-ABL1 rearrangements, in-frame fusions of EBF1-PDGFRB, BCR-JAK2 or STRN3-JAK2 and cryptic IGH-EPOR rearrangements.[63]
The recognition of this subgroup is of relevance, because of the poor
prognosis observed. Open issues are represented by difficulty in
detecting them with techniques other than gene expression profiling,
which is not routinely performed in all centers, and by the fact that
there is not a recurrent common lesion underlying the signature
identified. With this in mind, it is plausible that the use of TKIs
and/or mTOR inhibitors might be of benefit in these patients, as
suggested by xenograft models.[64,65]
Hypodiploid ALL, regarded as a poor prognosis group, has been extensively evaluated in pediatric ALL:[66] NGS proved that lesions involving receptor tyrosine kinases and RAS signaling (i.e. NRAS, KRAS, FLT3 and NF1)
can be detected in up to 70% of near haploid cases, whereas low
hypodiploid cases are characterized by lesions involving members of the
Ikaros family, particularly IKZF2, and by TP53
disruptions, that can be identified in 91.2% of these cases. In adult
ALL, these cases are characterized by nonrandom chromosomal losses and
the CDKN2A/B locus deletion as sole recurrent abnormality; as already reported in children, these cases frequently harbor TP53 mutations.[67]
TP53 disruption has been also recently evaluated in childhood and adult ALL. In children[68-71]
this is detected in 6.4% and 11.1% of relapsed B-ALL and T-ALL cases,
and, in a smaller minority of cases, also at diagnosis. A correlation
with poorer outcome has been shown. In adults, TP53
mutations are identified at diagnosis in 8.2% of cases (11.1% T-ALL and
6.4% B-ALL), and are preferentially identified in cases without
molecular aberrations, where they are detected in 14% of cases, and are
associated with refractoriness to chemotherapy.
Other lesions identified by NGS in B-lineage ALL, are represented by mutations in CREBBP and its paralogue, EP300 (p300),[72] which were identified in the relapse samples and appear to be more frequent in hyperdiploid relapsed cases.[73] Similarly, NT5C2
mutations, which confer increased enzymatic activity on the NT5C2
protein, which normally dephosphorylates nucleoside analogs, such as
mercaptopurine, used in consolidation and maintenance therapy, have
been described.[74] Results are summarized in Table 3.
T-lineage ALL:
In T-ALL, well-recognized aberrations include the T-cell receptor (TCR)
gene rearrangements, chromosomal deletions, and focal gene deletions (Table 4).[75-83]
Moreover, chromosomal rearrangements can also lead to in-frame fusion
genes encoding chimeric proteins with oncogenic properties such as PICALM-MLLT10, NUP214-ABL1 fusion formed on episomes, EML-ABL1, SET-NUP214 fusion and MLL gene rearrangements with numerous different partners. The prognostic significance of these lesions is uncertain.
Furthermore,
the ETP subgroup and/or myeloid-like subgroup emerged as a grey zone
between AML and T-ALL by applying genome-wide technologies.[18,84,85]
Initially, the reported incidence of this subgroup was established at
around 10% of T-ALL cases; however, with the better recognition of
these cases, its frequency is likely to be higher. Immunophenotype is
characterized by an early T-cell phenotype and co-expression of at
least one myeloid marker, while at the transcriptional level they have
a stem-cell like profile with overexpression of myeloid transcription
factors (including CEBPA, CEBPB, CEBPD),
and a set of micro-RNAs (miR-221, miR-222 and miR-223). NGS has
highlighted the presence of mutations usually found in acute myeloid
leukemia (IDH1, IDH2, DNMT3A, FLT3 and NRAS),[86] as well mutations in the ETV6 gene. Finally, these cases rarely harbor NOTCH1 mutations.[87] Overall, prognosis is poor in these cases.
A large set of mutations (Table 4) has been identified in T-ALL by re-sequencing and NGS: they include NOTCH1, FBW7, BCL11B, JAK1, PTPN2, IL7R and PHF6,
beyond those identified in ETPs; some of them have recognized
prognostic significance, whereas for others further studies are
required. In fact, NOTCH1 and/or FBW7 mutations,
which occur in more than 60% and about 20% of cases, respectively, are
usually associated with a favorable outcome. In the light of this, a
prognostic model has been recently proposed, defining as low-risk
patients those who harbor NOTCH1 and FBW7 mutations, and as high risk those without these mutations or with lesions involving RAS/PTEN.[83,88-91] At variance, JAK1
mutations, which increase JAK activity and alter proliferation and
survival have been associated with chemotherapy refractoriness and
should be considered as poor prognostic markers.[92-94]
Finally,
another group of mutations/lesions is possibly involved in
leukemogenesis, but their prognostic impact is either unknown or
absent. They include: 1) BCL11B lesions, which can induce a developmental arrest and aberrant self-renewal activity;[95,96] 2) PTPN2
- a negative regulator of tyrosine kinases-, mutations, often detected
in TLX1 overexpressing cases, T-ALL, NUP214-ABL+ patients and JAK1 mutated cases;[97,98] 3) mutations in IL7Ralpha, that lead to constitutive JAK1 and JAK3 activation and enhancement of cell cycle progression;[99,100] 4) PHF6 mutations;[101,102] 5) mutations in PTPRC, encoding the protein tyrosine phosphatase CD45, usually detected in combination with activating mutations of IL7R, JAK1 or LCK, and associated with downregulation of CD45 expression;[103] 6) mutations in CNOT3, presumed to be a tumor suppressor; 7) mutations of RPL5 and RPL10, which impair ribosomal activity.[104] Lastly, similarly to what is observed in relapsed B-ALL, NT5C2 mutations.[105]
Concluding Remarks
Due
to the reviewed evidence and the complexity of all the issues at play,
it is recommended that adult patients with ALL should be treated within
prospective clinical trials, which is the best way to ensure both
diagnostic accuracy and therapeutic efficacy. In the context of a
modern risk- and subset-oriented therapy, the early diagnostic work-up
is of the utmost importance and therefore needs to be carried out by
well trained and highly experienced personnel (Figure 4).
As a first step, it is mandatory to differentiate rapidly Ph+ from
Ph-ALL and to distinguish between major immunophenotypic subsets in the
latter group. The remaining diagnostic elements are available at a
later stage and permit a proper identification and treatment of the
several disease and risk entities. Ongoing research will permit the
further definition of novel subgroups with prognostic significance.
|
|
Figure 4. Diagnosis and subclassification of adult ALL.
To
confirm diagnosis and obtain clinically useful information, it is
necessary to 1) differentiate rapidly Ph-positive ALL from
Ph-negative ALL in order to allow an early introduction of
tyrosine kinase inhibitors in the former subset, 2) distinguish between
different clinico-prognostic Ph- ALL subsets, and 3) clarify diagnostic
issues related to the application of targeted therapy and risk-/minimal
residual disease (MRD)-oriented therapy. The early diagnostic phase
must be completed within 24-48 hours. Additional test for
cytogenetics/genetics, genomics and MRD rely on collection, storage and
analysis of large amounts of diagnostic material, and are usually
available at later time-points during therapy, however before taking a
decision for allogeneic stem cell transplantation (SCT). All this
requires a dedicated laboratory, and is best performed within a
prospective, well coordinated clinical trial.
|
References
- Vardiman
JW, Thiele J, Arber DA, et al. The 2008 revision of the World Health
Organization (WHO) classification of myeloid neoplasms and acute
leukemia: rationale and important changes. Blood 2009;114:937-951. http://dx.doi.org/10.1182/blood-2009-03-209262

- Bennett
JM, Catovsky D, Daniel MT, et al. French-American-British (FAB
Cooperative Group). Proposals for the classification of the acute
leukaemias. Br J Haematol 1976; 33:451-458. http://dx.doi.org/10.1111/j.1365-2141.1976.tb03563.x PMid:188440

- Bennett
JM, Catovsky D, Daniel MT, et al. The morphological classification of
acute lymphoblastic leukaemia: concordance among observers and clinical
correlations. Br J Haematol 1981; 47:553-561. http://dx.doi.org/10.1111/j.1365-2141.1981.tb02684.x PMid:6938236

- Jaffe
ES, Harris NL, Stein H, et al. Introduction an overview of the
classification of the lymphoid neoplasms. In: Swerdlow SH, Campo E,
Harris NL, et al, eds. WHO Classification of tumours of haematopoietic
and lymphoid tissue. IARC: Lyon
2008;158-166. PMid:18283093

- Lai
R, Hirsch-Ginsberg CF, Bueso-Ramos C. Pathologic diagnosis of acute
lymphocytic leukemia .Hematol Oncol Clin North Am.2000;14:1209-1235. http://dx.doi.org/10.1016/S0889-8588(05)70183-0

- d'Onofrio
G, Zini G, BainBJ (Translator). Morphology of Blood Disorders, 2nd
Edition. Wiley-Blackwell, 2014. ISBN: 978-1-118-44260)
- Sevilla DW, Colovai AI, Emmons FN, et al. Hematogones: a review and update. Leuk Lymphoma 2010;51:10-19. http://dx.doi.org/10.3109/10428190903370346 PMid:20001239

- Stein
P, Peiper S, Butler D, et al. Granular acute lymphoblastic leukemia. Am
J Clin Pathol 1983;80:545.PMid:6578676

- Gassmann
W, Löffler H, Thiel E, et al. Morphological and cytochemical findings
in 150 cases of T-lineage acute lymphoblastic leukaemia in adults.
German Multicentre ALL Study Group (GMALL). Br J Haematol 1997;
97:372-382. http://dx.doi.org/10.1046/j.1365-2141.1997.d01-2171.x PMid:9163604

- Bene
MC, Castoldi G, Knapp W, et al. Proposals for the immunological
classification of acute leukemias. European Group for the Immunological
Characterization of Leukemias (EGIL). Leukemia
1995;9:1783-1786. PMid:7564526

- Béné
MC, Nebe T, Bettelheim P, et al. Immunophenotyping of acute leukemia
and lymphoproliferative disorders: a consensus proposal of the European
LeukemiaNet Work Package 10. Leukemia 2011;25:567-574. http://dx.doi.org/10.1038/leu.2010.312 PMid:21252983

- Kalina
T, Flores-Montero J, van der Velden VH, et al. EuroFlow standardization
of flow cytometer instrument settings and immunophenotyping protocols.
Leukemia 2012;26:1986-2010. http://dx.doi.org/10.1038/leu.2012.122 PMid:22948490 PMCid:PMC3437409

- van
Dongen JJ, Lhermitte L, Böttcher S, et al. EuroFlow antibody panels for
standardized n-dimensional flow cytometric immunophenotyping of normal,
reactive and malignant leukocytes. Leukemia 2012;26:1908-1975. http://dx.doi.org/10.1038/leu.2012.120 PMid:22552007 PMCid:PMC3437410

- Coustan-Smith
E, Behm FG, Sanchez J, et al. Immunological detection of minimal
residual disease in children with acute lymphoblastic leukaemia..Lancet
1998;351:550-554. http://dx.doi.org/10.1016/S0140-6736(97)10295-1

- Janossy
G, Coustan-Smith E, Campana D. The reliability of cytoplasmic CD3 and
CD22 antigen expression in the immunodiagnosis of acute leukemia: a
study of 500 cases. Leukemia 1989;3:170-181 PMid:2465463

- Lenormand
B, Bene MC, Lesesve JF, et al. PreB1 (CD10-) acute lymphoblastic
leukemia: immunophenotypic and genomic characteristics, clinical
features and outcome in 38 adults and 26 children. The Groupe dEtude
Immunologique des Leucemies. Leuk Lymphoma
1998;28:329-342. PMid:9517504

- Geijtenbeek
TB, van Kooyk Y, van Vliet SJ, et al. High frequency of adhesion
defects in B-lineage acute lymphoblastic leukemia. Blood
1999;94:754-764. PMid:10397743

- Coustan-Smith
E, Mullighan CG, Onciu M, et al: Early T-cell precursor leukaemia: a
subtype of very high-risk acute lymphoblastic leukaemia. Lancet Oncol
2009;10:147-156. http://dx.doi.org/10.1016/S1470-2045(08)70314-0

- Vardiman
JW, Thiele J, Arber DA, et al. The 2008 revision of the World Health
Organization (WHO) classification of myeloid neoplasms and acute
leukemia: rationale and important changes. Blood 2009;114:937-951. http://dx.doi.org/10.1182/blood-2009-03-209262 PMid:19357394

- Matutes
E, Pickl WF, Van't Veer M, et al. Mixed-phenotype acute leukemia:
clinical and laboratory features and outcome in 100 patients defined
according to the WHO 2008 classification. Blood 2011;117:3163-171. http://dx.doi.org/10.1182/blood-2010-10-314682 PMid:21228332

- Paietta
E, Neuberg D, Richards S, et al. Rare adult acute lymphocytic leukemia
with CD56 expression in the ECOG experience shows unexpected phenotypic
and genotypic heterogeneity. Am J Hematol 2001;66:189-196. http://dx.doi.org/10.1002/1096-8652(200103)66:3<189::AID-AJH1043>3.0.CO;2-A

- Kantarjian
HM, Hirsch-Ginsberg C, Yee G, et al. Mixed-lineage leukemia revisited:
acute lymphocytic leukemia with myeloperoxidase-positive blasts by
electron microscopy. Blood
1990;76:808-813. PMid:2166608

- Hans
CP, Finn WG, Singleton TP, et al. Usefulness of anti-CD117 in the flow
cytometric analysis of acute leukemia. Am J Clin Pathol
2002;117:301-305. http://dx.doi.org/10.1309/RWCG-E5T9-GU95-LEWE PMid:11863227

- Faber
J, Kantarjian H, Roberts MW, Keating et al. Terminal deoxynucleotidyl
transferase-negative acute lymphoblastic leukemia. Arch Pathol Lab Med
2000;124:92-97 PMid:10629138

- Zhou
Y, Fan X, Routbort M, et al Absence of terminal deoxynucleotidyl
transferase expression identifies a subset of high-risk adult
T-lymphoblastic leukemia/lymphoma.Mod Pathol 2013;26:1338-1345. http://dx.doi.org/10.1038/modpathol.2013.78 PMid:23702731

- Pui
CH, Crist WM, Look AT. Biology and clinical significance of cytogenetic
abnormalities in childhood acute lymphoblastic leukemia. Blood
1990;76:1449-1463. PMid:2207320

- Secker-Walker
LM, Prentice HG, et al. Cytogenetics adds independent prognostic
information in adults with acute lymphoblastic leukaemia on MRC trial
UKALL XA. MRC Adult Leukaemia Working Party. Br J Haematol
1997;96:601-610. http://dx.doi.org/10.1046/j.1365-2141.1997.d01-2053.x PMid:9054669

- Group
Français de Cytogénétique Hématologique. Cytogenetic abnormalities in
adult acute lymphoblastic leukemia: correlations with hematologic
findings outcome. A Collaborative Study of the Group Français de
Cytogénétique Hématologique.. Blood 1996;88:3135-314.

- Wetzler
M, Dodge RK, Mrozek K, et al. Prospective karyotype analysis in adult
acute lymphoblastic leukemia: the cancer and leukemia Group B
experience. Blood 1999;93:3983-3993. PMid:10339508

- Kolomietz
E, Al-Maghrabi J, Brennan S, et al. Primary chromosomal rearrangements
of leukemia are frequently accompanied by extensive submicroscopic
deletions and may lead to altered prognosis. Blood 2001;97:3581-3588. http://dx.doi.org/10.1182/blood.V97.11.3581 PMid:11369654

- Moorman
AV, Harrison CJ, Buck GA, et al. Karyotype is an independent prognostic
factor in adult acute lymphoblastic leukemia (ALL): analysis of
cytogenetic data from patients treated on the Medical Research Council
(MRC) UKALLXII/Eastern Cooperative Oncology Group (ECOG) 2993 trial.
Blood 2007;109:3189-3197. http://dx.doi.org/10.1182/blood-2006-10-051912 PMid:17170120

- Pullarkat
V, Slovak ML, Kopecky KJ, Forman SJ, Appelbaum FR. Impact of
cytogenetics on the outcome of adult acute lymphoblastic leukemia:
results of Southwest Oncology Group 9400 study. Blood
2008;111:2563-2572. http://dx.doi.org/10.1182/blood-2007-10-116186 PMid:18156492 PMCid:PMC2254550

- Nahi
H, Hagglund H, Ahlgren T, et al. An investigation into whether
deletions in 9p reflect prognosis in adult precursor B-cell acute
lymphoblastic leukemia: a multi-center study of 381 patients.
Haematologica 2008;93:1734-1738. http://dx.doi.org/10.3324/haematol.13227 PMid:18728022

- Charrin
C, Thomas X, Ffrench M, et al. A report from the LALA-94 and LALA-SA
groups on hypodiploidy with 30 to 39 chromosomes and near-triploidy: 2
possible expressions of a sole entity conferring poor prognosis in
adult acute lymphoblastic leukemia (ALL). Blood 2004;104:2444-2451. http://dx.doi.org/10.1182/blood-2003-04-1299 PMid:15039281

- Felice
MS, Gallego MS, Alonso CN, et al. Prognostic impact of t(1;19)/
TCF3-PBX1 in childhood acute lymphoblastic leukemia in the context of
Berlin-Frankfurt-Münster-based protocols. Leuk Lymphoma
2011;52:1215-1221. http://dx.doi.org/10.3109/10428194.2011.565436 PMid:21534874

- Burmeister
T, Gökbuget N, Schwartz S, Fischer L, Hubert D, Sindram A, Hoelzer D,
Thiel E. Clinical features and prognostic implications of TCF3-PBX1 and
ETV6-RUNX1 in adult acute lymphoblastic leukemia. Haematologica
2010;95:241-246. http://dx.doi.org/10.3324/haematol.2009.011346 PMid:19713226
 PMCid:PMC2817026
PMCid:PMC2817026 
- Harrison
CJ, Moorman AV, Schwab C, et al. An international study of
intrachromosomal amplification of chromosome 21 (iAMP21): cytogenetic
characterization and outcome. Leukemia 2014;28:1015-1021 http://dx.doi.org/10.1038/leu.2013.317 PMid:24166298

- Moorman
AV, Schwab C, Ensor HM, et al. IGH@ translocations, CRLF2 deregulation,
and microdeletions in adolescents and adults with acute lymphoblastic
leukemia J Clin Oncol 2012; 30:3100-3108. http://dx.doi.org/10.1200/JCO.2011.40.3907 PMid:22851563

- Schardt
C, Ottmann OG, Hoelzer D, Ganser A. Acute lymphoblastic leukemia with
the (4;11) translocation: combined cytogenetic, immunological and
molecular genetic analyses. Leukemia
1992;6:370-374. PMid:1375695

- Faderl
S, Albitar M. Insights into the biologic and molecular abnormalities in
adult acute lymphocytic leukemia. Hematol Oncol Clin North Am.
2000;14:1267-1288. http://dx.doi.org/10.1016/S0889-8588(05)70186-6

- Westbrook
CA, Hooberman AL, Spino C, et al. Clinical significance of the BCR-ABL
fusion gene in adult acute lymphoblastic leukemia: a Cancer and
Leukemia Group B Study (8762). Blood.
1992;80:2983-2990 PMid:1467514

- Gleissner
B, Gökbuget N, Bartram CR, et al, German Multicenter Trials of Adult
Acute Lymphoblastic Leukemia Study Group. Leading prognostic relevance
of the BCR-ABL translocation in adult acute B-lineage lymphoblastic
leukemia: a prospective study of the German Multicenter Trial Group and
confirmed polymerase chain reaction analysis. Blood
2002;99:1536-1543. http://dx.doi.org/10.1182/blood.V99.5.1536 PMid:11861265

- Chiaretti
S, Vitale A, Cazzaniga G, et al.Clinico-biological features of 5202
patients with acute lymphoblastic leukemia enrolled in the Italian
AIEOP and GIMEMA protocols and stratified in age cohorts.
Haematologica. 2013;98:1702-1710. http://dx.doi.org/10.3324/haematol.2012.080432 PMid:23716539 PMCid:PMC3815170

- Rieder
H, Ludwig WD, Gassmann W, et al. Prognostic significance of additional
chromosome abnormalities in adult patients with Philadelphia chromosome
positive acute lymphoblastic leukaemia. Br J Haematol. 1996;95:678-691.
http://dx.doi.org/10.1046/j.1365-2141.1996.d01-1968.x PMid:8982045

- Ravandi
F, Jorgensen JL, Thomas DA, et al. Detection of MRD may predict the
outcome of patients with Philadelphia chromosome-positive ALL treated
with tyrosine kinase inhibitors plus chemotherapy. Blood. 2013 Aug
15;122:1214-1221. http://dx.doi.org/10.1182/blood-2012-11-466482 PMid:23836561 PMCid:PMC3976223

- Marks
DI, Moorman AV, Chilton L, et al. The clinical characteristics, therapy
and outcome of 85 adults with acute lymphoblastic leukemia and
t(4;11)(q21;q23)/MLL-AFF1 prospectively treated in the
UKALLXII/ECOG2993 trial. Haematologica. 2013;98:945-952. http://dx.doi.org/10.3324/haematol.2012.081877 PMid:23349309 PMCid:PMC3669452

- Schneider
NR, Carroll AJ, Shuster JJ, et al. New recurring cytogenetic
abnormalities and association of blast cell karyotypes with prognosis
in childhood T-cell acute lymphoblastic leukemia: a pediatric oncology
group report of 343 cases. Blood.
2000;96:2543-2549. PMid:11001909

- Lange
BJ, Raimondi SC, Heerema N, et al. Pediatric leukemia/lymphoma with
t(8;14)(q24;q11). Leukemia 1992;6:613-618. PMid:1385638

- Parolini
M, Mecucci C, Matteucci C, et al. Highly aggressive T-cell acute
lymphoblastic leukemia with t(8;14)(q24;q11): extensive genetic
characterization and achievement of early molecular remission and
long-term survival in an adult patient. Blood Cancer J. 2014 Jan
17;4:e176. http://dx.doi.org/10.1038/bcj.2013.72 PMid:24442205 PMCid:PMC3913941

- Martinelli
G, Iacobucci I, Storlazzi CT, et al: IKZF1 (Ikaros) deletions in
BCRABL1-positive acute lymphoblastic leukemia are associated with short
disease-free survival and high rate of cumulative incidence of relapse:
a GIMEMA AL WP report. J Clin Oncol 2009; 27:5202-5207. http://dx.doi.org/10.1200/JCO.2008.21.6408 PMid:19770381

- Mullighan
CG, Su X, Zhang J, et al: Children's Oncology Group: Deletion of IKZF1
and prognosis in acute lymphoblastic leukemia. N Engl J Med 2009;
360:470-480. http://dx.doi.org/10.1056/NEJMoa0808253 PMid:19129520 PMCid:PMC2674612

- van
der Veer A, Zaliova M, Mottadelli F, et al. IKZF1 status as a
prognostic feature in BCR-ABL1-positive childhood ALL. Blood 2014;
123:1691-8. http://dx.doi.org/10.1182/blood-2013-06-509794 PMid:24366361

- Kuiper
RP, Waanders E, van der Velden VH, et al: IKZF1 deletions predict
relapse in uniformly treated pediatric precursor B-ALL. Leukemia 2010;
24:1258-1264. http://dx.doi.org/10.1038/leu.2010.87 PMid:20445578

- Moorman
AV, Schwab C, Ensor HM, et al. IGH@ translocations, CRLF2 deregulation,
and microdeletions in adolescents and adults with acute lymphoblastic
leukemia J Clin Oncol 2012; 30:3100-3108. http://dx.doi.org/10.1200/JCO.2011.40.3907 PMid:22851563

- van
der Veer A, Waanders E, Pieters R, et al. Independent prognostic value
of BCR-ABL1-like signature and IKZF1 deletion, but not high CRLF2
expression, in children with B-cell precursor ALL. Blood
2013;122:2622-2629. http://dx.doi.org/10.1182/blood-2012-10-462358 PMid:23974192 PMCid:PMC3795461

- Mullighan
CG, Collins-Underwood JR, Phillips LA, et al: Rearrangement of CRLF2 in
B-progenitor- and Down syndrome-associated acute lymphoblastic
leukemia. Nat Genet 2009; 41:1243-1246. http://dx.doi.org/10.1038/ng.469 PMid:19838194 PMCid:PMC2783810

- Yoda
A, Yoda Y, Chiaretti S, et al. Yoda A, Yoda Y, Chiaretti S, et al:
Functional screening identifies CRLF2 in precursor B-cell acute
lymphoblastic leukemia. Proc Natl Acad Sci U S A 2010; 107:252-257. http://dx.doi.org/10.1073/pnas.0911726107 PMid:20018760 PMCid:PMC2806782

- Hertzberg
L, Vendramini E, Ganmore I, et al: Down syndrome acute lymphoblastic
leukemia, a highly heterogeneous disease in which aberrant expression
of CRLF2 is associated with mutated JAK2: a report from the
International BFM Study Group. Blood 2010; 115:1006-1117. http://dx.doi.org/10.1182/blood-2009-08-235408 PMid:19965641

- Haferlach
T, Kohlmann A, Schnittger S, et al: Global approach to the diagnosis of
leukemia using gene expression profiling. Blood 2005; 106:1189-1198. http://dx.doi.org/10.1182/blood-2004-12-4938 PMid:15878973

- Chiaretti
S, Li X, Gentleman R, et al. Gene expression profiles of B-lineage
adult acute lymphocytic leukemia reveal genetic patterns that identify
lineage derivation and distinct mechanisms of transformation. Clin
Cancer Res. 2005;11:7209-7219. http://dx.doi.org/10.1158/1078-0432.CCR-04-2165 PMid:16243790

- Den
Boer ML, van Slegtenhorst M, De Menezes RX, et al: A subtype of
childhood acute lymphoblastic leukaemia with poor treatment outcome: a
genome-wide classification study. Lancet Oncol 2009; 10:125-134. http://dx.doi.org/10.1016/S1470-2045(08)70339-5

- Harvey
RC, Mullighan CG, Wang X, et al: Identification of novel cluster groups
in pediatric high-risk B-precursor acute lymphoblastic leukemia with
gene expression profiling: correlation with genome-wide DNA copy number
alterations, clinical characteristics, and outcome. Blood
2010; 116:4874-4884. http://dx.doi.org/10.1182/blood-2009-08-239681 PMid:20699438 PMCid:PMC3321747

- Roberts
KG, Morin RD, Zhang J, et al. Genetic alterations activating kinase and
cytokine receptor signaling in high-risk acute lymphoblastic leukemia.
Cancer Cell 2012;22:153-166. http://dx.doi.org/10.1016/j.ccr.2012.06.005 PMid:22897847 PMCid:PMC3422513

- Maude
SL, Tasian SK, Vincent T, et al. Targeting JAK1/2 and mTOR in murine
xenograft models of Ph-like acute lymphoblastic leukemia. Blood
2012;120:3510-3518. http://dx.doi.org/10.1182/blood-2012-03-415448 PMid:22955920 PMCid:PMC3482861

- Roberts
KG, Li Y, Payne-Turner D, et al. Targetable kinase-activating lesions
in Ph-like acute lymphoblastic leukemia. N Engl J Med
2014;371:1005-1015. PMid:25207766

- Holmfeldt
L, Wei L, Diaz-Flores E, et al. The genomic landscape of hypodiploid
acute lymphoblastic leukemia. Nat Genet 2013;45:242-252. http://dx.doi.org/10.1038/ng.2532 PMid:23334668 PMCid:PMC3919793

- Mühlbacher
V, Zenger M, Schnittger S, et al. Acute lymphoblastic leukemia with low
hypodiploid/near triploid karyotype is a specific clinical entity and
exhibits a very high TP53 mutation frequency of 93%. Genes Chromosomes
Cancer 2014;53:524-536. http://dx.doi.org/10.1002/gcc.22163 PMid:24619868

- Hof
J, Krentz S, van Schewick, et al: Mutations and deletions of the TP53
gene predict nonresponse to treatment and poor outcome in first relapse
of childhood acute lymphoblastic leukemia. J Clin Oncol 2011;
29:3185-93. http://dx.doi.org/10.1200/JCO.2011.34.8144 PMid:21747090

- Krentz
S, Hof J, Mendioroz A, et al: Prognostic value of genetic alterations
in children with first bone marrow relapse of childhood B-cell
precursor acute lymphoblastic leukemia. Leukemia 2013;27:295-304. http://dx.doi.org/10.1038/leu.2012.155 PMid:22699455

- Chiaretti
S, Brugnoletti F, Tavolaro S, et al: TP53 mutations are frequent in
adult acute lymphoblastic leukemia cases negative for recurrent fusion
genes and correlate with poor response to induction therapy.
Haematologica 2013;98:e59-61. http://dx.doi.org/10.3324/haematol.2012.076786 PMid:23403321 PMCid:PMC3640132

- Stengel
A, Schnittger S, Weissmann S, et al. TP53 mutations occur in 15.7% of
ALL and are associated with MYC-rearrangement, low hypodiploidy and a
poor prognosis. Blood 2014;124:251-258. http://dx.doi.org/10.1182/blood-2014-02-558833 PMid:24829203

- Mullighan CG, Zhang J, Kasper LH, et al: CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature 2011; 471:235-239. http://dx.doi.org/10.1038/nature09727 PMid:21390130 PMCid:PMC3076610

- Inthal
A, Zeitlhofer P, Zeginigg M, et al. CREBB HAT domain mutations prevail
in relapse cases of high hyperdiploid childhood acute lymphoblastic
leukemia. Leukemia. 2012;26:1797-1803. http://dx.doi.org/10.1038/leu.2012.60 PMid:22388726 PMCid:PMC4194312

- Meyer
JA, Wang J, Hogan LE, et al. Relapse-specific mutations in NT5C2 in
childhood acute lymphoblastic leukemia. Nat Genet 2013; 45:290-294. http://dx.doi.org/10.1038/ng.2558 PMid:23377183 PMCid:PMC3681285

- Barber
KE, Martineau M, Harewood L, et al. Amplification of the ABL gene in
T-cell acute lymphoblastic leukemia. Leukemia 2004;18:1153-1156. http://dx.doi.org/10.1038/sj.leu.2403357 PMid:15057249

- Ferrando
AA, Neuberg DS, Dodge RK, et al. Prognostic importance of TLX1 (HOX11)
oncogene expression in adults with T-cell acute lymphoblastic
leukaemia. Lancet 2004;363:535-536. http://dx.doi.org/10.1016/S0140-6736(04)15542-6

- Ballerini
P, Busson M, Fasola S, et al. NUP214-ABL1 amplification in
t(5;14)/HOX11L2-positive ALL present with several forms and may have a
prognostic significance. Leukemia 2005;19:468-470. http://dx.doi.org/10.1038/sj.leu.2403654 PMid:15674415

- Asnafi
V, Buzyn A, Thomas X, et al. Impact of TCR status and genotype on
outcome in adult T-cell acute lymphoblastic leukemia: a LALA-94 study.
Blood 2005;105:3072-3078. http://dx.doi.org/10.1182/blood-2004-09-3666 PMid:15637138

- Burmeister
T, Gokbuget N, Reinhardt R, Rieder H, Hoelzer D, Schwartz S.
NUP214-ABL1 in adult T-ALL: the GMALL study group experience. Blood
2006;108:3556-3559. http://dx.doi.org/10.1182/blood-2006-04-014514 PMid:16873673

- Baldus
CD, Burmeister T, Martus P, et al. High expression of the ETS
transcription factor ERG predicts adverse outcome in acute
T-lymphoblastic leukemia in adults. J Clin Oncol. 2006;24:4714-4720. http://dx.doi.org/10.1200/JCO.2006.06.1580 PMid:16954520

- Baldus
CD, Martus P, Burmeister T, et al. Low ERG and BAALC expression
identifies a new subgroup of adult acute T-lymphoblastic leukemia with
a highly favorable outcome. J Clin Oncol. 2007;25:3739-3745. http://dx.doi.org/10.1200/JCO.2007.11.5253 PMid:17646667

- Bergeron
J, Clappier E, Radford I, et al. Prognostic and oncogenic relevance of
TLX1/HOX11 expression level in T-ALLs. Blood. 2007;110:2324-2330. http://dx.doi.org/10.1182/blood-2007-04-079988 PMid:17609427

- Asnafi
V, Buzyn A, Le Noir S, et al: NOTCH1/FBXW7 mutation identifies a large
subgroup with favourable outcome in adult T-cell acute lymphoblastic
leukemia (TALL): a GRAALL study. Blood 2009; 113:3918-3924 http://dx.doi.org/10.1182/blood-2008-10-184069 PMid:19109228

- Chiaretti
S, Messina M, Tavolaro S, et al: Gene expression profiling identifies a
subset of adult T-cell acute lymphoblastic leukemia with myeloid-like
gene features and over-expression of miR-223. Haematologica 2010;
95:1114-1121. http://dx.doi.org/10.3324/haematol.2009.015099 PMid:20418243 PMCid:PMC2895035

- Coskun
E, Neumann M, Schlee C, et al. MicroRNA profiling reveals aberrant
microRNA expression in adult ETP-ALL and functional studies implicate a
role for miR-222 in acute leukemia. Leuk Res 2013;37:647-56. http://dx.doi.org/10.1016/j.leukres.2013.02.019 PMid:23522449

- Zhang
J, Ding L, Holmfeldt L, Wu G, et al: The genetic basis of early T-cell
precursor acute lymphoblastic leukaemia. Nature 2012; 481:157-163. http://dx.doi.org/10.1038/nature10725 PMid:22237106 PMCid:PMC3267575

- Van
Vlierberghe P, Ambesi-Impiombato A, Perez-Garcia A, et al: ETV6
mutations in early immature human T cell leukemias. J Exp Med 2011;
208:2571-2579. http://dx.doi.org/10.1084/jem.20112239 PMid:22162831 PMCid:PMC3244026

- Park
MJ, Tak T, Oda M, et al: FBXW7 and NOTCH1 mutations in childhood T cell
acute lymphoblastic leukaemia and T cell non-Hodgkin lymphoma. Br J
Haematol 2009; 145:198-206. http://dx.doi.org/10.1111/j.1365-2141.2009.07607.x PMid:19245433

- Mansour
MR, Sulis ML, Duke V, et al: Prognostic implications of NOTCH1 and
FBXW7 mutations in adults with T-cell acute lymphoblastic leukemia
treated on the MRC UKALLXII/ECOG E2993 protocol. J Clin Oncol 2009;
27:4352-4356. http://dx.doi.org/10.1200/JCO.2009.22.0996 PMid:19635999 PMCid:PMC2744275

- Ben
Abdelali R, Asnafi V, Leguay T, et al: Group for Research on Adult
Acute Lymphoblastic Leukemia: Pediatric-inspired intensified therapy of
adult T-ALL reveals the favorable outcome of NOTCH1/FBXW7 mutations,
but not of low ERG/BAALC expression: a GRAALL study. Blood 2011;
118:5099-5107. http://dx.doi.org/10.1182/blood-2011-02-334219 PMid:21835957

- Trinquand
A, Tanguy-Schmidt A, Ben Abdelali R, et al. Toward a
NOTCH1/FBXW7/RAS/PTEN-based oncogenetic risk classification of adult
T-cell acute lymphoblastic leukemia: a Group for Research in Adult
Acute Lymphoblastic Leukemia study. J Clin Oncol. 2013;31:4333-4342. http://dx.doi.org/10.1200/JCO.2012.48.5292 PMid:24166518

- Flex
E, Petrangeli V, Stella L, et al: Somatically acquired Jak1 mutation in
adult acute lymphoblastic leukemia. J Exp Med 2008;205:751-758. http://dx.doi.org/10.1084/jem.20072182 PMid:18362173 PMCid:PMC2292215

- Jeong
EG, Kim MS, Nam HK, et al: Somatic mutations of JAK1 and JAK3 in acute
leukemias and solid cancers. Clin Cancer Res 2008; 14:3716-3721. http://dx.doi.org/10.1158/1078-0432.CCR-07-4839 PMid:18559588

- Asnafi
V, Le Noir S, Lhermitte L, et al: JAK1 mutations are not frequent
events in adult T-ALL: a GRAALL study. Br J Haematol 2010;148:178-179. http://dx.doi.org/10.1111/j.1365-2141.2009.07912.x PMid:19764985

- Gutierrez
A, Kentsis A, Sanda T, et al: The BCL11B tumor suppressor is mutated
across the major molecular subtypes of T-cell acute lymphoblastic
leukemia. Blood 2011; 118:4169-4173. http://dx.doi.org/10.1182/blood-2010-11-318873 PMid:21878675 PMCid:PMC3204734

- Kraszewska
MD, Dawidowska M, Szczepanski T, et al: T-cell acute lymphoblastic
leukaemia: recent molecular biology findings. Br J Haematol 2012;
156:303-315. http://dx.doi.org/10.1111/j.1365-2141.2011.08957.x PMid:22145858

- Kleppe
M, Lahortiga I, El Chaar T, et al: Deletion of the protein tyrosine
phosphatase gene PTPN2 in T-cell acute lymphoblastic leukemia. Nat Gen
2010; 42:530-535. http://dx.doi.org/10.1038/ng.587 PMid:20473312 PMCid:PMC2957655

- Kleppe
M, Soulier J, Asnafi V, et al: PTPN2 negatively regulates oncogenic
JAK1 in T-cell acute lymphoblastic leukemia. Blood 2011; 117:7090-7098.
http://dx.doi.org/10.1182/blood-2010-10-314286 PMid:21551237

- Shochat
C, Tal N, Bandapalli OR, et al. Gain-of-function mutations in
interleukin-7 receptor-a (IL7R) in childhood acute lymphoblastic
leukemias. J Exp Med. 2011;208:901-908. http://dx.doi.org/10.1084/jem.20110580 PMid:21536738 PMCid:PMC3092356

- Zenatti
PP, Ribeiro D, Li W, et al: Oncogenic IL7R gain-of-function mutations
in childhood T-cell acute lymphoblastic leukemia. Nat Genet 2011;
43:932-939. http://dx.doi.org/10.1038/ng.924 PMid:21892159

- Van
Vlierberghe P, Palomero T, Khiabanian H, et al: PHF6 mutations in
T-cell acute lymphoblastic leukemia.Nat Genet 2010; 42:338-342 http://dx.doi.org/10.1038/ng.542 PMid:20228800 PMCid:PMC2847364

- Wang
Q, Qiu H, Jiang H, et al: Mutations of PHF6 are associated with
mutations of NOTCH1, JAK1 and rearrangement of SET-NUP214 in T-cell
acute lymphoblastic leukemia. Haematologica 2011; 96:1808-1814. http://dx.doi.org/10.3324/haematol.2011.043083 PMid:21880637 PMCid:PMC3232263

- Porcu
M, Kleppe M, Gianfelici V, et al: Mutation of the receptor tyrosine
phosphatase PTPRC (CD45) in T-cell acute lymphoblastic leukemia. Blood
2012; 119:4476-4479. http://dx.doi.org/10.1182/blood-2011-09-379958 PMid:22438252

- De
Keersmaecker K, Atak ZK, Li N, et al: Exome sequencing identifies
mutation in CNOT3 and ribosomal genes RPL5 and RPL10 in T-cell acute
lymphoblastic leukemia. Nat Genet 2013;45:186-190. http://dx.doi.org/10.1038/ng.2508 PMid:23263491

- Tzoneva
G, Perez-Garcia A, Carpenter Z, et al: Activating mutations in the
NT5C2 nucleotidase gene drive chemotherapy resistance in relapsed ALL.
Nat Med 2013;19:368-371. http://dx.doi.org/10.1038/nm.3078 PMid:23377281 PMCid:PMC3594483

[TOP]




































 PMCid:PMC2817026
PMCid:PMC2817026 




































































