Received: December 2, 2015
Accepted: September 16,2016
Mediterr J Hematol Infect Dis 2016, 8(1): e2016047, DOI 10.4084/MJHID.2016.047
This article is available on PDF format at:

Sara Martinelli, Antonio Cuneo, Luca Formigaro, Maurizio Cavallari, Enrico Lista, Francesca Maria Quaglia, Maria Ciccone, Antonella Bardi, Eleonora Volta, Elisa Tammiso, Elena Saccenti, Olga Sofritti, Giulia Daghia, Massimo Negrini, Melissa Dabusti, Paolo Tomasi, Sabrina Moretti, Francesco Cavazzini and Gian Matteo Rigolin
Section of Hematology, Azienda Ospedaliero-Universitaria S.Anna, Ferrara, Italy.

| This is an Open Access article distributed
under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |
|
Abstract Chronic lymphocytic leukemia (CLL)
displays an extremely variable clinical behaviour. Accurate
prognostication and prediction of response to treatment are important
in an era of effective first-line regimens and novel molecules for high
risk patients. Because a plethora of prognostic biomarkers were
identified, but few of them were validated by multivariable analysis in
comprehensive prospective studies, we applied in this survey stringent
criteria to select papers from the literature in order to identify the
most reproducible prognostic/predictive markers. Each biomarker was
analysed in terms of reproducibility across the different studies with
respect to its impact on time to first treatment (TTFT), progression
free survival (PFS), overall survival (OS) and response to treatment.
We were able to identify the following biomarkers as the most reliable
in guiding risk stratification in the daily clinical practice:
17p-/TP53 mutations, IGHV unmutated configuration, short telomeres and
11q-. However, the method for measuring telomere length was not
validated yet and 11q- was predictive of inferior OS only in those
patients who did not receive FCR-like combinations. Stage and
lymphocytosis were predictive of shorter TTFT and age, high serum
thymidine kinase levels and poor performance status were predictive of
shorter OS. Using our criteria no parameter was found to independently
predict for inferior response to treatment. |
Introduction
Chronic lymphocytic leukemia (CLL) displays a variable clinical behaviour, with many patients living for years without symptoms and other patients requiring early therapeutic intervention attaining short lasting responses and succumbing to their disease in a few years. Therefore, survival in this chronic lymphoproliferative disorder largely depends on the rapidity of disease progression and on the quality and duration of response to treatment. The availability of effective first-line regimens[1-6] makes prognostication and prediction of response to treatment an important exercise in clinical practice, especially in young and/or fit patients who may benefit of aggressive regimens including allogeneic bone marrow transplantation.[7,8] Clinical staging is a simple measure of disease burden and still represents a convenient, yet insufficient means of assessing prognosis, because it does not identify those patients with limited disease who have a high probability to progress, and it does not predict the quality and duration of response to treatment. A plethora of biomarkers have been identified in the last decades which may predict disease outcome,[9] but few of them were validated in the context of prospective studies using adequate statistic considerations to weigh the risk of each parameter by multivariable analysis. Meanwhile, our understanding of CLL biology greatly improved providing a basis for a better understanding of the biologic role of prognostic markers.[10] The pathogenesis of CLL is the result of a complex interplay between i) lymphocytes carrying a restricted repertoire of BCR,[11] ii) the mutational status of the variable portion of the immunoglobulin heavy chain (IGHV) gene determining different behaviour of neoplastic lymphocyte in response to antigen stimulation, iii) cell activation and interaction with the microenvironment,[12] iv) genetic lesions[13] (Figure 1). Each of these fundamental mechanisms is associated with specific biomarkers that identify high-risk CLL, as summarized in Figure 2. Central to this categorization of prognostic markers is the concept that antigenic stimulation of neoplastic lymphocytes with a restricted set of BCR may promote inhibition of apoptosis, survival and proliferation within the lymph node and bone marrow microenvironment, with consequent multiple cycles of cell division, telomere shortening and genetic instability, with disease-host interactions ultimately shaping variable clinical phenotypes (Figure 2).[14]
In this review clinicobiologic features predicting outcome are discussed in correlation with their pathogenic role and applicability in clinical practice.
| Figure 2. Pathogenic steps and corresponding prognostic markers. |
Eligibility Criteria and Literature Search
Based on previous analyses that identified clinical and biologic characteristics having prognostic significance,[9,10,15-17] the following 18 biomarkers were included in this survey: stereotyped receptors and BCR subsets, CD38, ZAP70, CD49d, IGHV gene mutational status, 17p-/TP53 mutations, 11q-, telomere length, complex karyotype, NOTCH1 and SF3B1 mutations, age, gender, performance status, stage, lymphocytosis, beta-2-microglobulin, thymidine kinase.
A literature search was then performed to identify studies on the prognostic value of these biomarkers in CLL. We searched PubMed to identify all citations from January 2000 to April 2016 describing the role of the selected parameters in predicting the outcome for newly diagnosed CLL patients. The search was performed using a combination of MeSH controlled vocabulary and text words. The following terms were used: “Leukemia, Lymphocytic, Chronic, B-Cell"[Mesh], "Prognosis"[Mesh], "Clinical Trial" [Publication Type], "Receptors, Antigen, B-Cell"[Mesh], CD38, ZAP70, CD49d, IGHV, IGVH, 17p[All Fields], TP53[All Fields], 11q[All Fields], "Telomere"[Mesh], telomere, complex karyotype, NOTCH1, SF3B1, "beta 2-Microglobulin"[Mesh], thymidine kinase.
Only full length publications satisfying the following requirements were included in the review: i) English language; ii) at least 100 patients included; iii) multivariate analysis including salient clinical data and genetic testing (IGHV mutational status, 17p deletion and 11q deletion); iv) prospective design of the study (clinical trial) or single/multicentre study using a learning cohort and a validation cohort or consecutive series; v) at least one endpoint being time to first treatment (TTFT), progression free survival (PFS), overall survival (OS), overall response rate (ORR) or complete response (CR) rate. Manuscripts describing the prognostic impact of the selected parameters in the context of patients starting unconventional or experimental treatment were not included, as well as studies including patients with monoclonal B-cell lymphocytosis.
The search criteria identified 3,845 citations. After duplicate removal and evaluation of all remaining manuscripts, 27 papers met the criteria for inclusion in this study. The characteristics and salient data of these papers are presented in Table 1.
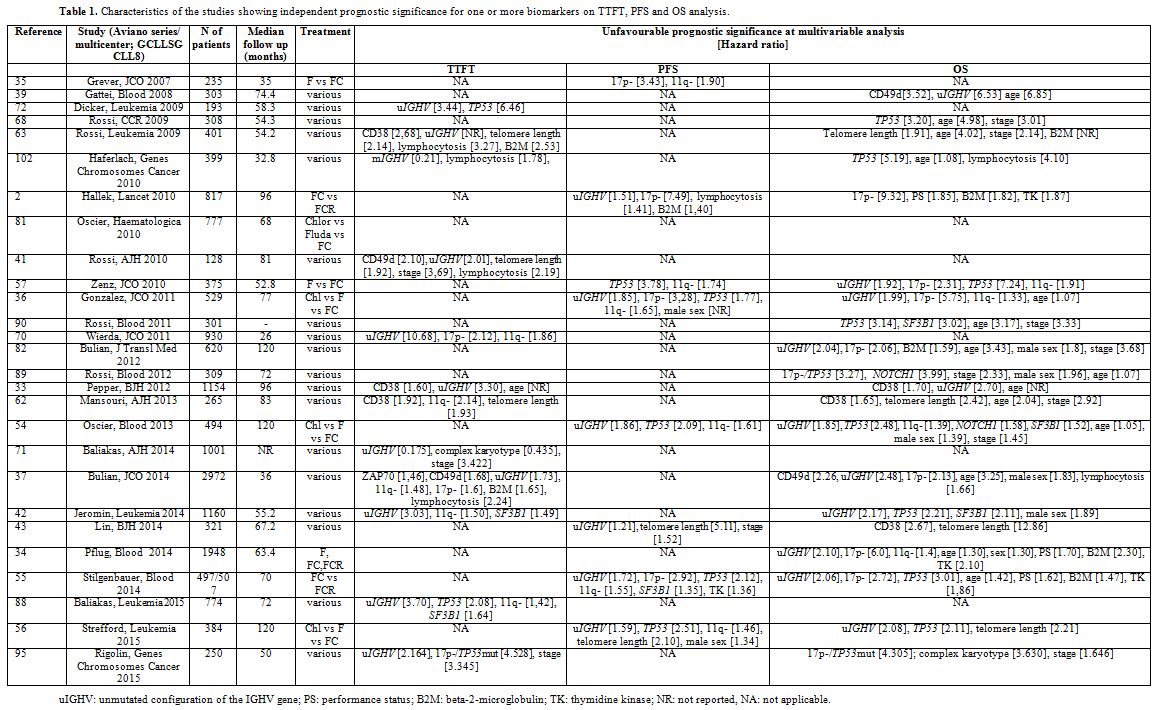 |
Table 1. Characteristics of the studies showing independent prognostic significance for one or more biomarkers on TTFT, PFS and OS analysis. |
Results
Predictors of outcome (TTFT, PFS and OS). Figure 3 represents,
for each parameter, the total number of studies analyzing its
prognostic impact and the number of studies in which the parameter
showed independent prognostic significance in terms of TTFT, PFS and/or
OS. Hazard ratios for each marker are reported in Table 1.
| Figure 3. Number of studies assessing the impact of each parameter in terms of a) TTFT; b) PFS; c) OS: the blue part of each column represents the number of studies showing independent negative impact on prognosis (“significant”); the red part represents the number ofstudies showing no prognostic impact (“not significant”). |
1) BCR repertoire and stereotyped receptors
Pathogenic role:
CLL lymphocytes express a restricted set of BCR due to non-random usage
of gene families coding for the variable portion of the Ig.[11]
Furthermore, some CLL cases express highly homogeneous sequences of the
heavy chain complementarity determining region 3 (HCDR3), a phenomenon
referred to as “stereotyped” BCR[18] that was shown to occur in up to
one-third of the cases.[19-21]
The similarity of the BCR from
various CLL patients suggests that the precursors of B-CLL cells were
chosen for their antigen-binding capabilities by antigen(s) of
restricted nature and structure.
Prognostic impact:
Biased BCR usage and stereotyped receptors did not meet the criteria
adopted in this review. The prognostic significance of stereotyped BCR
was only assessed in studies that did not include a comprehensive
assessment of salient genetic parameters, and no prospective clinical
trial was designed including determination of BCR stereotypy in the
diagnostic workup. CLL with stereotyped BCR showed shorter TTFT in a
study including genetic parameters;[22] however no multivariable
analysis was performed in this analysis.
2) Interaction with the microenvironment and activation markers
Pathogenic role:
The interaction between neoplastic lymphocytes responding to BCR
stimulation and the microenvironment plays a fundamental role in CLL
pathogenesis.[23] As a consequence, the natural history of the disease
is dictated, in part, by lymphocyte survival and growth in the lymph
nodes that favours the accumulation of genomic alterations, especially
within proliferation centres and/or CD38-positive cells.[24,25]
Biomarkers reflecting the capability to respond to BCR engagement or to
adhere to niches in the bone marrow or lymph node (i.e. CD38, ZAP70,
CD49d, lipoprotein lipase, TCL1 expression) were correlated with
progressive disease[26-30] and, to the contrary, a signature of anergic
lymphocyte that poorly responds to BCR stimulation was observed in
indolent CLL.[31,32]
Prognostic impact:
The threshold for CD38-positivity varied across studies. In a
comprehensive analysis of 1154 early stage CLL seen at 4 European
centres,[33] CD38+ was predictive of a shorter TTFT (median 9.3 years
vs not reached) and shorter survival (median 14,7 years) by
multivariable analysis (Table 1).
Interestingly a more rapid disease progression requiring treatment was
observed in CD38 positive CLL in patients with “favourable” genetic
profile.[25] However, the prognostic significance of CD38 was overcome
by more robust genetic parameters in several clinical trials, as shown
in Figure 3.[34-36]
ZAP-70
was predictive of a shorter TTFT in only one study,[37] and no impact
was shown on PFS or OS in all studies using a complete assessment of
prognostic parameters when the multivariable analysis was
applied.[35,38-43] One study showed an independent impact of ZAP70 on
OS;[44] however, it was not considered in this survey as it used an
unusually low expression level (>10%) as a positivity cut-off and
because the detection method is not reproducible.[9]
CD49d
expression was predictive of shorter interval between diagnosis and
disease progression, with a median TTFT of approximately 4 years in
CD49d-positive cases.[38,44] Its predictive value was confirmed by
multivariable analysis in two analyses[37,41] and an independent
adverse impact on OS was documented in two studies using heterogeneous
treatment.[37,39]
No study with comprehensive assessment of
prognostic markers was performed to assess the independent prognostic
impact of lipoproteine lipase and TCL1 overexpression.
3) IGHV mutational status and genetic features
Pathogenic role: Neoplastic lymphocytes carrying ≤2% mutations of the IGHV
gene compared to the nearest germline sequence are referred to as
“unmutated” CLL. The lymphocytes in this CLL subset respond to antigen
stimulation by activating intracellular signalling and entering the G1
phase of the cell cycle more efficiently than IGHV “mutated” CLL.[23,45,46] Consequently neoplastic lymphocyte carrying unmutated IGHV
sequence, i) undergo more cell divisions in vivo as shown by
incorporation of deuterated water[12] and, ii) carry shorter telomeres
and accumulate more genomic defects[47] than lymphocytes with
mutated IGHV.
The
molecular pathogenesis of CLL is centred around some lesions. i.e. TCL1
or miR-29 overexpression and/or miR15a-16 deletion, producing the
disease in the animal model.[48-50] These lesions are associated with a
number of chromosomes and genetic aberrations which may appear soon
during clonal expansion (13q-, +12, MYD88, NOTCH1 mutations), or later following disease progression or selection by treatment (11q-, 17p-, mutations of TP53, ATM, SF3B1, BIRC3).[51,52] Disruption of the TP53 and ATM
pathway is associated with resistance to DNA damaging agents and
genetic instability leading to the emergence of subclones accounting
for disease progression.[9]
Prognostic impact:
- Unmutated IGHV sequences: Ever since the first reports,[26,53] unmutated IGHV
proved to be a robust unfavourable prognostic marker.[16] In a
prospective study by Shanafelt and coworkers[38] TTFT in 1004 CLL was
2,8 years in unmutated CLL as compared to 11 years in mutated CLL. In
our analysis, a significantly shorter TTFT with very high hazard ratios
(Table 1) was observed in 8
prospective series, 4 of which had enrolled more than 700 cases. PFS
was shorter in IGHV unmutated CLL in 2 large studies using fludarabine
and cyclofosfamide (FC) or fludarabine, cyclofosfamide and rituximab
(FCR)2 or chlorambucil (Chlor), fludarabine (F) or FC[36] and the
observed difference held when including assessment of new gene
mutations (i.e. SF3B1 and NOTCH1) and telomere length in the multivariable model.[54-56] In the CLL4 trial of the GCLLSG using F or FC the IGHV
configuration in 294 patients showed no independent impact on PFS[57]
as was the case with a US Intergroup Phase III Trial E2997 that
assessed the IGHV mutational status in 195 patients;[35] however these
2 analyses were numerically smaller than the other studies.
Interestingly, a shorter survival was noted in IGHV unmutated CLL in a number of large studies (Table 1, Figure 3)
with 85,1% five-year OS in unmutated CLL vs 91.4% five-year OS in
mutated CLL in a recent analysis that pooled data form 3 randomized
studies of the GCLLSG.[34]
- Telomeres:
Telomeres are specific non-coding nucleotide sequences consisting of
6–12 kbp of TTAGGG-repeats located at the ends of eukaryotic
chromosomes that are necessary for the complete replication and
stability of the chromosome. Because they are eroded upon each cell
division, their length reflects the replicative history of a cell.[58]
In CLL cells telomeres are shorter that in normal B-lymphocytes[59] and
those patients with telomere length below the median observed value
were found to frequently carry unmutated IGHV
gene[60] and unfavourable prognosis.[61] In our analysis, a
significantly shorter TTFT was found in CLL with shortened telomeres in
3 studies,[41,62,63] 2 of which had partially overlapping cohorts (Table 1).
Short telomeres were independently associated with inferior PFS in two
large studies assessing the most significant prognostic
parameters[43,56] and negatively impacted on OS in 4 studies that
included patients treated with various regimens(Table 1, Figure 3).[43,56,62,63]
- 17p-/TP53 mutations and 11q deletion: These aberrations alter cell-cycle and DNA-repair pathways. In the case of a functional TP53
pathway, DNA damage activates p53 through the activation of ATM, thus
inducing cell cycle arrest through p21 and apoptosis. 17p13 deletion
causes loss of one TP53
allele and determines resistance to DNA-damaging drugs through
haploinsufficiency.[64] 17p13 deletion is associated in the vast
majority of cases with inactivating mutations of the remaining TP53 allele. The TP53
mutation may occur independent of 17p deletion and may inactivate p53
function by a dominant negative effect or by duplication of the
chromosome segment containing the mutated TP53 gene, a genetic rearrangement referred to as uniparental disomy.[65] Disruption of the TP53
pathway by deletion and/or mutation leads to resistance to apoptosis
and genetic instability and indeed 17p- patients frequently exhibit
complex chromosome defects and multiple genetic lesions.[55,66] The
negative impact of 17p13 deletion on OS and clinical response to
fludarabine were clearly documented as early as 1995,[67] and a similar
negative impact was shown to be associated with TP53 mutations,
independent of the presence of 17p13 deletion.[36,57,68] Interestingly,
minor clones carrying TP53
mutations (<20% of the cells) were equally shown by sensitive next
generation sequencing techniques to predict for an inferior
outcome.[69] The papers selected for this review show that the
aberrations leading to disruption of the TP53 pathway, either by 17p deletion or by inactivating TP53 gene mutation, or both, have a deleterious impact on all outcome measure, as shown in Table 1 and Figure 3.
A hazard ratio of 3,96 of being treated after 26 months vs. patients
with 13q- as single aberration was recorded in a large prospective
study of 930 patients[70] and more rapid disease progression requiring
treatment was observed in one large multicentre study[71] and in one
single centre study.[72] At variance, one large analysis conducted at a
referral laboratory receiving samples from several institutions did not
find an impact on TTFT for TP53 mutations.[42] Even though the presence
of mutated IGHV gene and early stage reduced the adverse effect of this genetic aberration in a minority of patients,[73,74] 17p-/TP53
mutations definitely identify patients with dismal outcome with the
current treatment regimens using alkylating agents, purine analogues,
and anti CD20 monoclonal antibodies. Indeed shorter PFS and OS were
uniformly reported in all the studies (Table 1, Figure 3). Interestingly, TP53
disruption appears to be associated with more frequent progression in
the relapsed/refractory setting under novel active BCR-targeted
therapies.[75-77] 11q22-23 deletion characterizes a CLL subtype
with extensive nodal involvement and inferior prognosis.[78] The
minimal region of deletion involves the ataxia-teleangiectasia mutated (ATM) gene, with concurrent mutation of the remaining ATM
allele occurring in 30-40% of 11q- cases and causing extreme telomere
shortening[79] and dismal prognosis.[80] Because ATM is a very large
gene, mutational studies were not performed in clinical trials and
papers included in this review refer to patients with 11q-. TTFT and
PFS were shorter in patients with 11q- in the majority of studies (Table 1, Figure 3).
It is noteworthy, however, that the negative impact on PFS was overcome
by adding rituximab to FC.[2] Likewise, some reports demonstrated an
adverse impact of 11q- on OS,[81] whereas more studies using effective
regimens based on purine analogues and alkylating agents with
rituximab[2,55,82] did not detect any difference by multivariable
analysis. Thus, it appears that the negative prognostic impact of 11q-
is abated in young and/or fit patients treated by modern
chemoimmunotherapy regimens.
- Lesions involving NOTCH1 and SF3B1: Mutations causing activation of the NOTCH1 pathway, with consequent activation of non-canonical NFkB signalling, may promote cell survival and resistance to apoptosis.[83-85] SF3B1
mutations cause altered splicing of a number of targets including
FOXP1, that encodes for a forkhead transcription machinery,[86]
promoting resistance to fludarabine-based treatment through as yet
unknown mechanisms.[87] Shorter TTFT was noted in patients with SF3B1 mutations in 2 studies,[42,88] whereas NOTCH1 mutation did not predict for more rapid disease evolution (Table 1). Likewise, NOTCH1 mutations did not impact on PFS, whereas shorter PFS was observed in SF3B1 mutated
patients treated by FC or FCR in the CLL8 study.[55] This correlation
was not observed in the UKCLL4 study using chlorambucil, or fludarabine
with or without cyclophosphamide (Table 1).[54] Rossi et al. showed an adverse impact on OS of NOTCH1 mutation.[89] OS was shorter in SF3B1 and NOTCH1 mutated patients in the UKCLL4 trial[54] and not in the CLL8 trial (Table 1).[55] It is noteworthy that two more studies showed an independent impact of the SF3B1 mutation on overall survival (Figure 3).[42,90]
- Conventional banding analysis:
Metaphase karyotyping represented the first biomarker having prognostic
significance in CLL, in the seminal paper by Juliusson et al..[91] More
recently complex karyotype was included in a prognostic scoring system
predictive of time to first treatment and overall survival.[92] The
presence of chromosome aberrations was predictive of an inferior
outcome in those patients without detectable aberrations by
fluorescence in situ hybridization (FISH).[93] Unbalanced chromosome
translocations, frequently occurring in the context of complex
karyotype, were shown to be independent prognostic factors in a
study[94] and complex karyotype predicted for a shorter TTFT[71] and OS[95]
by multivariable analysis. The prognostic value of complex
karyotype was also demonstrated in relapsed/refractory CLL patients
treated with ibrutinib-based regimens, where this parameter showed a
stronger impact on the outcome than del(17p).[96] After data for this
review had been collected, a comprehensive prospective study on 161 CLL
patients with relevant comorbidity showed the independent prognostic
role of complex karyotype for survival following chlorambucil-based
chemoimmunotherapy.[97] Thus, there is mounting evidence that
chromosome banding analysis with novel efficient mitogens may
substantially contribute to the identification of CLL patients with
adverse prognosis.
4) Disease/ host characteristics
Pathogenic role:
Markers of tumor burden and proliferative activity of neoplastic cells
may have an obvious impact on prognosis. Clinical stage, peripheral
lymphocytosis, bone marrow infiltration pattern, serum markers such as
soluble CD23 and ß2-microglobulin, an extracellular protein associated
with the class I major histocompatibility complex, represented for many
years valuable prognostic markers.[98] Likewise, markers of
proliferative activity, i.e. lymphocyte doubling time and serum
thymidine kinase-1 (TK1), a cellular enzyme involved in the DNA
synthesis in the G1/S phase of the cell cycle and reflecting the number
of dividing neoplastic cells, were shown to have a significant
prognostic impact.
Age and gender may have an evident impact on
prognosis. According to the global health observatory data repository
of the WHO (available at the link http://www.who.int/gho/mortality_burden_disease/life_tables/situation_trends/en/)
life expectancy in 2012 at the age of 60 was 19 and 23 years in Europe
for males and females respectively (21 and 24 years in the Americas).
Given the availability of treatments that are able to modify the
natural history of the disease and to prolong survival,[99] host
characteristics such as performance status and comorbidities may also
have a significant role as prognosticators.[34,55]
Prognostic impact:
The standardization of chemiluminescence immunoassay for the assessment
of serum thymidine kinase-1 (TK) levels significantly facilitated the
introduction of this marker into clinical practice,[100] and
age-related normal reference values were recently defined and
validated.[101] When this test was included among diagnostic workup
within prospective trials, raised TK levels (i.e. ≥10 U/L) were
predictive of shorter PFS[55] and survival in the GCLLSG trials.[34]
Likewise, serum beta-2-microglobulin levels proved an independent
prognostic parameter on more outcome measures in several studies (Table 1, Figure 3).
Lymphocytosis,
ECOG performance status, stage and gender showed a variable impact on
prognosis. Direct quantitation of the disease burden such lymphocytosis
and stage are predictive of shorter TTFT and have a variable impact on
other outcome measures,[102] however, they lacked significant
prognostic value in the largest analysis based on pooled data from
several GCLLSG studies.[34]
Age and PS had no impact on TTFT and PFS, whereas they showed significant impact on OS (Table 1, Figure 3).
Factors predictive of response to treatment
No
paper with the stringent characteristics adopted in our analysis was
found to address the impact of biomarkers on predicting response
treatment, with two notable exceptions:
a) 17p-/TP53 mutations and 11q-.
There is evidence that carrying 17p-/TP53 mutation conveys a low probability to achieve a clinical response with chemoimmunotherapy.[67] ORR was lower in 17p-/TP53
mutated patients in 3 large randomized trials comparing FC vs FCR,[2] F
vs FC[57] and Chlorambucil vs F vs FC.[81] Even though these data were
not validated by multivariable analysis, the probability to attain a
meaningful response was very low for the 17p- group in the CLL8 trial,
with a CR rate of 0% and 5% and an ORR rate of 51,6% and 75% with FC
and FCR, respectively.[2,55] Interestingly, TP53 mutations were shown to represent an independent predictive factor of shorter time to chemorefractoriness.[68]
b) Lack of efficacy of anti CD20 monoclonal antibodies rituximab and ofatumumab in NOTCH1 mutated patients.
Recent
evidence was provided that response to chemoimmunotherapy containing
anti-CD20 monoclonal antibodies may be negatively influenced by the
presence of NOTCH1 mutations.
The adjunct of ofatumumab to chlorambucil significantly improved PFS in
the total population of a phase III study;[103] while not influencing
response to treatment, NOTCH1
mutation was associated with shorter PFS in the ofatumamuab plus
chlorambucil arm (17.7 months vs. 23.3, HR 1.86 p=0.01), making the
addition of the monoclonal antibody to chlorambucil irrelevant in terms
of PFS over chlorambucil alone.[103] In the CLL8 study[55] there was no
significant difference in terms of ORR in the FC arm and in the FCR arm
depending on the NOTCH1 mutation status; however, while the association of rituximab with FC improved ORR in patients with wild type NOTCH1 (88,1 vs. 96,6%; p<0.01), no difference in ORR was noted in NOTCH1 mutated patients in the FC and FCR arms (87,1% vs. 90%). PFS was superimposable in NOTCH1 mutated patients in the FCR (34,2 months) and FC arm (33,9 months). Taken together these data suggest that NOTCH1
mutation is a predictive marker for reduced benefit from the addition
of rituximab or ofatumumab to chemotherapy. Interestingly, CLL cases
carrying NOTCH1 mutations are
characterized by low CD20 expression levels deriving at least in part
from histone deacetylase-dependent transcriptional repression, an
observation that may explain the low sensitivity of these patients to
anti-CD20 monoclonal antibodies.[104]
Conclusion
Using stringent criteria we were able to identify 16 parameters, i.e. CD38, CD49d, unmutated IGHV, 17p-/TP53 mutations, 11q-, telomere length, complex karyotype, NOTCH1 and SF3B1 mutations, age, gender, performance status (PS), stage, lymphocytosis, beta-2-microglobulin, thymidine kinase, with unfavourable prognostic significance on TTFT, PFS and/or OS by multivariable analysis in prospective clinical trials or in the context of well-organized studies. The observed TTFT, PFS, and OS for each of these markers in the corresponding studies is shown in Figure 4.
| Figure 4. Time to first treatment (a), PFS (b) and OS (c) in the presence of the unfavourable biomarker (reference number at the bottom ofeach column). Chl: chlorambucil; F: fludarabine; FC: fludarabine and cyclophosphamide, FCR for FC plus rituximab. |
Since 17p-/TP53 mutations, unmutated IGHV gene configuration and 11q- proved to be independent predictors of outcome in at least 2/3 studies (Figure 3)
and their detection methods were standardized,[105-107] these markers
are to be considered the most reliable for usage in clinical practice.
Telomere length proved a reproducible predictor of the outcome but the
detection method is not standardized yet, even if a recent study
confirmed the reproducibility of results obtained with monochrome
multiplex Q-PCR (MMQ-PCR) and single telomere length analysis (STELA),
opening the way for the assay standardisation.[56] Complex karyotype as
detected by stimulation with novel mitogens[66] may represent a novel
predictor of unfavourable outcome.[71,95-97]
While simple measures
of disease burden such as stage and lymphocytosis do not play a role
any more as prognostic markers, host characteristics such as poor PS
and advanced age still play a relevant role in predicting OS.
It
is noteworthy that evolution of treatment may overcome the significance
of some of these prognostic factors; thus, while maintaining its
predictive value on OS in patients not eligible to modern
chemoimmunotherapy regimens, 11q- lost its unfavourable significance in
those patients treated by FCR.[34,55] Importantly, mechanism-based
treatment in the refractory/relapsed setting showed high efficacy in
“high risk” patients;[75] thus the introduction of new oral agents
targeting kinase signalling or BCL2 will likely change the significance
and role of many of these markers.[108]
References











































































































[TOP]