Sehnaz
Alp1 and Murat Akova2
1 Associate
Professor, Hacettepe University, Faculty of Medicine, Department of
Infectious Diseases and Clinical Microbiology, Ankara, Turkey
2
Professor, Hacettepe University, Faculty of Medicine, Department of
Infectious Diseases and Clinical Microbiology, Ankara, Turkey
Corresponding
author: Murat Akova. Professor, Hacettepe University, Faculty of
Medicine, Department of Infectious Diseases and Clinical Microbiology,
Ankara, Turkey. E-mail:
akova.murat@gmail.com
Published: January 1, 2017
Received: July 27, 2016
Accepted: November 11, 2016
Mediterr J Hematol Infect Dis 2017, 9(1): e2017002 DOI
10.4084/MJHID.2017.002
This article is available on PDF format at:

This is an Open Access article distributed
under the terms of the Creative Commons Attribution License
|
|
Abstract
Recipients of
hematopoietic stem cell transplantation (HSCT) are at
substantial risk of bacterial, fungal, viral, and parasitic infections
depending on the time elapsed since transplantation, presence of
graft-versus-host disease (GVHD), and the degree of immunosuppression.
Infectious complications in HSCT recipients are associated with high
morbidity
and mortality. Bacterial infections constitute the major cause of
infectious
complications, especially in the early post-transplant period. The
emergence of
antibacterial resistance complicates the management of bacterial
infections in
this patient group. Multidrug-resistant bacterial infections in this
group of
patients have attracted considerable interest and may lead to
significant
morbidity and mortality. Empirical antibacterial therapy in patients
with HSCT
and febrile neutropenia has a critical role for survival and should be
based on
local epidemiology. This review attempts to provide an overview of risk
factors
and epidemiology of emerging resistant bacterial infections and their
management in HSCT recipients.
|
Introduction
Hematopoietic
stem cell transplantation (HSCT) has become the treatment of choice to
cure or improve the outcomes of a wide variety of haematological
malignancies and disorders.[1-4]
HSCT can be performed
by the transfer of hematopoietic stem cells from the donor to the
recipient (allogeneic HSCT) or by the return of previously harvested
cells of the same individual (autologous HSCT) after administration of
conditioning regimens.[4]
Myeloablative (MA)
conditioning leads to profound pancytopenia, and also breaks down
mucosal barriers, which might result in seeding of residing
microorganisms of the gastrointestinal system into the bloodstream.
Therefore, infectious complications begin to appear in the early
post-transplant period. Nonmyeloablative (NMA) conditioning has the
advantages of reduced regimen-related toxicity and transplant-related
mortality. Therefore, patients being referred for HSCT but not eligible
to receive a myeloablative conditioning may have the opportunity to
benefit from HSCT. Recipients of NMA allogeneic HSCT experience a
heterogeneous duration and degree of pancytopenia according to the
administered regimen. NMA regimens with lower mucosal toxicity and
myelosuppression provide a low incidence of infectious complications
within the early period after transplantation. Immune recovery after
NMA regimens was shown to be faster than that was seen following MA
regimens, and improved immune reconstitution was associated with lower
incidence of life-threatening infectious complications. Even though
myelosuppressive potential of NMA regimens seems to be milder than MA
regimens, the severity and duration of lymphodepletion is assumed to be
similar, because of the implementation of immunosuppressive treatment
to prevent graft rejection.[4-8]
Risk factors for bacterial and
resistant bacterial infections in patients with HSCT
Infectious
complications are the major contributors of morbidity and mortality,
especially within one year following HSCT. In the early post-transplant
period, presence of neutropenia and mucosal damage predispose patients
to infections. Presence and severity of graft-versus-host disease
(GVHD) and immunosuppressive treatment for it have a considerable
impact on the degree of overall immunosuppression and risk of
infection.[4,7]
The frequent use of
central venous catheters brings about a substantial risk for severe,
often recurrent, and potentially lethal infections.[9-11]
Recipient factors such as age, comorbidities, and previous exposure to
infectious agents prior to transplant, and the type of transplant, due
to the distinct duration required for immune reconstitution, also
influence the risk of infectious complications.[4]
Initiating
broad-spectrum empirical antibacterial therapy results in decreased
mortality in febrile neutropenic HSCT recipients. On the other hand,
the use of such therapy has the risk of selection of resistant
pathogens.[9,12-14]
Fluoroquinolone prophylaxis in haematology settings led emerging
fluoroquinolone resistance.[15-20]
This prophylaxis has also been associated with emerging
methicillin-resistant Staphylococcus
aureus (MRSA), multidrug-resistant (MDR) Escherichia coli,
and Pseudomonas
aeruginosa bacteraemia, and Clostridium difficile
infections.[21-25]
Consequently, empirical carbapenem use in patients receiving quinolone
prophylaxis has increased, a practice may, in turn, result in increased
carbapenem-resistant bacterial infections.[16,26]
In addition, prolonged and/or repeated hospitalisations, intensive care
unit (ICU) stay, severity of illness, healthcare-associated infections,
presence of urinary catheter and older age are considered as major risk
factors for resistant bacterial infections.[12,26-30] Main risk factors for certain
resistant bacterial infections are summarised in the Table 1.
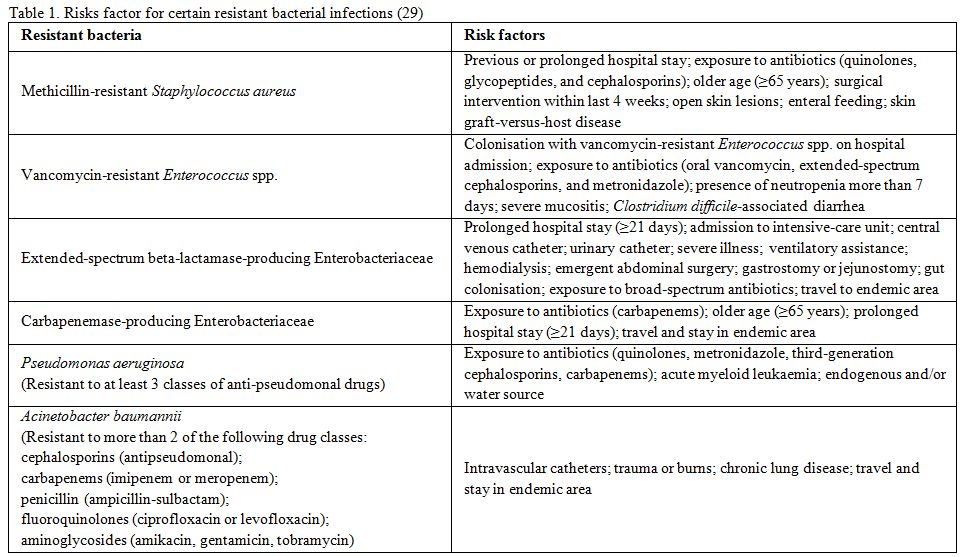 |
Table
1. Risks factor for certain resistant bacterial infections[29] |
Antibacterial
resistance in patients with HSCT
The
data on epidemiology of bacterial infections and their resistance
patterns in HSCT recipients mostly reflect isolates from bloodstream
infections (BSIs) which are the most frequent microbiologically
documented bacterial infections. The rate of BSIs varies between 20-30%
of allogeneic and 5% of autologous HSCT recipients, especially within
pre-engraftment phase. Even though bacterial pneumonia and skin and
soft tissue infections are also common among these patients, microbial
aetiology may remain undocumented.[29,31]
During
1960s and 1970s, the incidence of gram-negative infections was high in
haematology settings. Nevertheless, the incidence of gram-positive
pathogens increased during mid-1980s and 1990s as a result of extensive
use of indwelling catheters, early-generation fluoroquinolone
prophylaxis and broad-spectrum empirical anti-gram-negative
antibacterial therapy.[12,29,32-34]
Afterwards, coagulase-negative staphylococci were reported as the most
common bacterial etiologic agents isolated from blood cultures in most
centres.[10,35]
However, recent
reports from a number of centres revealed drug-resistant gram-negative
pathogens such as ESBL-producing gram-negative bacteria, multidrug
resistant (MDR) P.
aeruginosa, Acinetobacter
baumannii, Stenotrophomonas
maltophilia, and carbapenemase-producing gram-negative
bacteria as the causative agents of increasing numbers of infections.[9,12,36-44]
In countries where high rates of antibiotic resistance exist,
ESBL-producing or MDR gram-negative bacteria contribute up to 13-14% of
clinical isolates.[26,28,40,45]
A significant increase in the prevalence of resistant gram-positive
cocci such as MRSA and vancomycin-resistant enterococci (VRE) have also
been reported and stated as the overriding resistant pathogens in some
centres.[46,47]
Penicillin-resistant viridans
streptococci and penicillin-resistant Streptococcus pneumoniae (PRSP)
are less common, yet they may be the causative agents of severe
infections.[9,10,12,48]
The
epidemiology of bacterial infections and their resistance patterns show
distinct geographic and inter-centre variability. Being aware of the
current data on local epidemiology of predominant pathogens and close
monitoring of their resistance patterns are of great importance,
especially in empirical antibacterial treatment decisions.[12,29,49,50]
Recent
reviews on epidemiology of BSIs in cancer patients, primarily with
hematologic malignancies including HSCT recipients, revealed that among
all BSI isolates, coagulase-negative staphylococci and
Enterobacteriaceae (frequently
E. coli) were the most common pathogens followed by P. aeruginosa, S. aureus,
viridans streptococci, and enterococci. The approximate rates of these
commonly encountered pathogens were: 25% (range: 5-60%) for
coagulase-negative staphylococci; 25% (range: 6-54%) for
Enterobacteriaceae; 10% (range: 0-30%) for P. aeruginosa; 6%
(range: 0-20%) for S.
aureus; 5% (range: 0-16%) for viridans streptococci; and
5% (range: 0-38%) for enterococci.[29,49,51]
A
brief information on the epidemiology of global resistance data for
gram-positive and gram-negative bacteria is given below in each
corresponding title. An online website showing the current drug
resistance rates and antimicrobial use worldwide is also available at ‘http://resistancemap.cddep.org’.
Gram-Negative Bacteria
E. coli is one of
the most frequent pathogens causing bacteraemia in patients with cancer
and neutropenia.[49,51-53]
Production of one or more extended spectrum beta-lactamases (ESBLs) is
the main resistance mechanism against broad-spectrum penicillins and
cephalosporins in enteric gram-negative pathogens. Many ESBL-producing E. coli are also
resistant to non-beta-lactam antibiotics including aminoglycosides and
quinolones with altered resistance mechanisms.[52-54]
ESBL-encoding plasmids may also encode resistance to aminoglycosides,
tetracyclines, sulphonamides and trimethoprim.[52,55]
These plasmids frequently encode an inhibitor-resistant beta-lactamase,
which confers resistance to beta-lactam-beta-lactamase inhibitor
combinations including amoxicillin-clavulanate and
piperacillin-tazobactam.[52,55,56] Aminoglycoside resistance among E. coli
and other gram-negative enteric pathogens is determined by
aminoglycoside-modifying enzymes which can be encoded on the same
plasmid with ESBLs.[52] E. coli was the
second most frequent carbapenem-resistant Enterobacteriaceae (CRE)
following Klebsiella
pneumoniae. In a recent US survey, the incidence of CRE
was determined as 2.93 per 100.000 population.[52,57]
One
of the most significant carbapenemases described in Enterobacteriaceae
is New Delhi metallo-beta-lactamase-1 (NDM-1). This enzyme is prevalent
in the Indian subcontinent, but also frequently reported in Balkans and
the Middle East.[52,58] The
bacteria harbouring this enzyme have spread worldwide and are usually
only susceptible to colistin, tigecycline and fosfomycin, although
susceptibility to these agents is not universal.[52,59] Since E. coli
infections are very frequent in the outpatient settings, it is feared
that a progressive increase in the prevalence of NDM-1 producing E. coli may occur.[52,58]
Plasmid-mediated colistin resistance (via mcr-1 colistin
resistance gene) has recently been described in E. coli isolates
worldwide from mainly livestock and less frequently in human samples.[52,60-65]
The implications of this finding may be horrendous since the offending
plasmid can easily be transferred between E. coli strains and
to K. pneumonia
and P. aeruginosa.[52,66]
As a matter of fact, recent reports already noted the presence of this
gene from plasmids in Salmonella
and K. pneumoniae.[52,67-70]
Along
with ESBLs as the main resistance mechanism to broad-spectrum
penicillins and cephalosporins in enteric gram-negative pathogens,
carbapenem resistance has become the most important epidemiologic and
therapeutic challenge in K.
pneumoniae.[52,58]
There are mainly 3 classes of carbapenemases involved including KPC
(Class A), OXA-48 (Class D) and NDM (Class B) for which different
epidemiological reservoirs exist.[52,58,59,71-74]
A specific KPC-2 or KPC-3-producing clone has been widely disseminated
worldwide contributing the spread of resistance.[52,58]
Carbapenem-resistant isolates usually show MDR pattern and are
susceptible only to colistin, fosfomycin and tigecycline. However,
there is also emergence of resistance against these antibiotics.[52,75,76]
P. aeruginosa
strains with high resistance rates to aminoglycosides, ceftazidime,
quinolones, piperacillin-tazobactam and carbapenems are usually
reported from Southern and Eastern part of Europe.[52,77]
Several beta-lactamases have been described for causing resistance and
these include AmpC, ESBL (particularly PER-1) and
metallo-beta-lactamases.[52,55] Carbapenem resistance in P. aeruginosa is
mostly due to porin deficiencies and rarely caused by carbapenemase
production.[52,78]
Emergence of colistin resistance in P. aeruginosa has
also been reported.[52,79]
The most frequent Class A ESBLs found in A. baumannii
are PER-, GES- and VEB-type enzymes. These beta-lactamases confer
resistance to extended-spectrum cephalosporins, but inhibited by
tazobactam and clavulanic acid.[52,78] TEM-, SHV- and CTX-M-type ESBLs are
rarely found in A.
baumannii. Class B beta-lactamases (metalloenzymes) are
also reported in A.
baumannii
and include IMP-, VIM- and NDM-type enzymes. These beta-lactamases
provide activity against not only to carbapenems, but also to
broad-spectrum cephalosporins and penicillins.[52,80] Class D, OXA-type carbapenemases
are the most widespread carbapenemases in A. baumannii.[52,55]
These enzymes cause weak resistance to carbapenems. Thus, high-level
resistance usually require other mechanisms involved such as efflux and
porin loss.[52,78]
The ArmA enzyme is the most frequent methylase which is responsible for
high-level resistance to all aminoglycosides in A. baumannii. The
gene responsible for this enzyme is often identified among
OXA-23-producing A.
baumannii strains. Other methylases are also described.[52,78]
Overexpression of efflux pumps can provide resistance to quinolones.
These pumps also use aminoglycosides, tetracyclines, chloramphenicol
and trimethoprim as substrates. Thus, quinolone resistance can be
selected by non-quinolone antibiotics as well. Usually several of these
mechanisms are present in MDR Acinetobacter
isolates.[52]
For treatment of MDR gram-negative infections, especially due to
carbapenem-resistant Enterobacteriaceae, Pseudomonas
species, and Acinetobacter
species, colistin (polymyxin E) has been increasingly used as a
therapeutic option, administered as monotherapy or in combination
regimens, even though limited data exist on its use in haematology
patients and HSCT recipients.[26-28,81,82] There are many reports on
successful combination regimens for MDR gram-negative infections.[26,83-85]
Colistin plus rifampicin treatment has shown in vitro and in vivo
synergistic activity for A.
baumannii infections.[86,87]
However, in a multicentre, randomized clinical trial, colistin plus
rifampicin revealed no difference in infection-related mortality and
length of hospital stay in the treatment of serious infections due to
extensively drug-resistant A.
baumannii
as compared to colistin alone, but a significant increase in
microbiological eradication rate was determined in the colistin plus
rifampicin arm.[88] In a recent
study, survival
benefit with combination therapy (colistin plus carbapenem or
tigecycline plus carbapenem) was demonstrated in patients with
KPC-producing K.
pneumoniae bacteraemia.[89]
Gram-Positive Bacteria
Methicillin
resistance is the hallmark of antimicrobial resistance in S. aureus and
coagulase-negative staphylococci.[52]
While, vancomycin has long been successfully used for treatment of MRSA
infections, emergence of S.
aureus strains with vancomycin MICs ≥2 mg/L has coincided
with reports of treatment failures.[9,90] Community-acquired MRSA (CA-MRSA)
infections have emerged as a global problem since the beginning of the
21st
century.[51,90-92]
Although CA-MRSA strains initially caused mainly skin and soft tissue
infections in healthy individuals and some certain populations such as
homeless and imprisoned people, increased rates of bacteraemia both in
community and hospital setting; ventilator associated pneumonia; and
surgical site infections have recently been reported.[52,94,95]
CA-MRSA isolates usually remain susceptible to many non-beta-lactam
antibiotics including clindamycin and trimethoprim-sulfamethoxazole
(TMP-SMX).[52,96]
Coagulase-negative
staphylococci are the most common cause of nosocomial BSIs and are
responsible almost one-third of all healthcare-associated bacteraemia.
The incidence is highest in those with cancer and neutropenia and those
with catheter- and/or prosthetic device-related infections.[29,52,97] Multiple antibiotic resistance is
highly encountered among hospital isolates and usually related with
methicillin resistance.[52,98] Resistance to vancomycin is very
rare, however a 20.8% resistance to teicoplanin was reported from UK,
particularly in S.
haemolyticus.[98]
Penicillin-resistant
pneumococci are more likely to show higher resistance to other classes
of antimicrobials. Current figures of resistance in the US include 35%
to macrolides, 10% to clindamycin, 30% to TMP-SMX, 18% to doxycycline
and 2% to respiratory quinolones.[52,99] Higher rates of macrolide
resistance are reported from Europe.[52,100]
Viridans
streptococci can cause infective endocarditis, especially in patients
with compromised heart valves, and they can also produce bacteraemia
and septic shock particularly in patients with neutropenia.[51,52,101]
Although these bacteria are susceptible to most antimicrobials,
beta-lactam resistance, due to the altered penicillin binding proteins
has emerged and may cause a significant problem especially in patients
with immunosuppression and bacteraemia.[51,52,102]
Ceftriaxone and cefepime resistance has been reported up to 23 and 25%,
respectively in strains isolated from hospitalised or cancer patients.[52,103,104]
Vancomycin is highly effective on such strains.[52]
Among
all enterococci, Enterococcus faecium is the most challenging one in
terms of antibacterial resistance and therapy. In the US, enterococci
are the second most common bacteria isolated from catheter-related
(CR)-BSIs.[52,105]
Enterococci are
intrinsically resistant to many antimicrobials, but also easily acquire
mutations and exogenous genes to develop further resistance.[52,106]
While aminopenicillin resistance is rare in E. faecalis, it is
encountered around 90% of nosocomial E. faecium
isolates.[52,100,106]
Beta-lactamase production is infrequently associated with resistance
and can be overcome with the use of beta-lactamase inhibitor compounds.
The production of PBP5 with low affinity to penicillins is the major
culprit for beta-lactam resistance.[52,106]
High-level resistance to all aminoglycosides eliminates the synergistic
activity of penicillins and vancomycin both of which can enhance
activity of aminoglycosides in enterococci with low-to-moderate
resistance. High-level aminoglycoside resistance has increased
in
both E. faecalis
and E. faecium during
the last 3 decades.[52,100] Glycopeptide resistance in
enterococci is a much bigger problem in the US than in Europe and
elsewhere. By 2007, >80% of E.
faecium isolates
in the US hospitals were reported to be resistant to vancomycin whereas
in Europe only Ireland reported a resistance rate of >50%.[52,100,106,107] Similarly, MDR enterococci is
much more prevalent in the US. [52,106]
Enterococci are the third most frequent agents of bacteraemia in
haematological cancer patients and HSCT recipients and may affect up to
12% of all transplant patients. On these patient groups, a shift from E. faecalis to E. faecium has
resulted in higher rates of VRE infections.[51,52]
However, similar to the general epidemiology, VRE infections constitute
a less significant problem in Western European transplant centres with
<5% of enterococci being resistant to vancomycin.[52,104]
Resistance to linezolid and daptomycin is rarely reported.[52,108]
Newer
agents with activity against glycopeptide non-susceptible gram-positive
pathogens, such as daptomycin, linezolid, and tigecycline are being
increasingly used in various clinical settings.[9,12,27-29,109]
One of the major drawbacks of daptomycin is the inactivation of the
drug by pulmonary surfactant, which limits its use in treatment of
pneumonia. Moreover, treatment failure in staphylococcal central
nervous system infection was noticed.[26,110]
Even though daptomycin had not been evaluated in controlled trials in
haematology patients, its efficacy on gram-positive infections in
neutropenic patients has been reported.[26,111,112]
The clinical utility of tigecycline is limited by its low peak-serum
concentrations, and increased failure and mortality rates.[26,113,114]
C. difficile infection
(CDI) is among the major concerns in patients undergoing HSCT. Risk
factors for CDI in HSCT patients are specified as exposure to
broad-spectrum antimicrobial agents, receipt of chemotherapy prior to
conditioning for HSCT, total body irradiation, presence of acute GVHD,
and VRE colonisation.[115-118]
The outcomes of CDI
include increased morbidity and mortality due to increased risk of
developing complications such as colitis or toxic megacolon, extended
hospital stays, necessity to discontinue the required antibiotics, and
increased healthcare costs.[119,120]
The emergence
of an epidemic strain termed as ‘North American PFGE type 1 or NAP1’ is
associated with large outbreaks in Europe and the United States. NAP1
has a genetic alteration that results in enhanced toxin production and
has been associated with increased severity of CDI, higher relapse and
mortality rates.[9,121,122]
Even
though the studies conducted in 1980s and 1990s revealed that orally
administered metronidazole and vancomycin showed equal effectiveness
for treatment of CDI,[9,123,124]
with the emergence of the epidemic strain, reports of higher rates of
treatment failure or delayed treatment responses have appeared with
metronidazole as compared to oral vancomycin.[125-128]
There are variable data on the outcomes of CDI in haematology settings,
but treatment response to metronidazole and vancomycin is reported to
be similar.[26,129-131]
While
initiation of treatment for CDI, age, white blood cell count, and serum
creatinine level should be taken into consideration as indicators for
severe or complicated course.[122,132]
For the initial episode of mild-to-moderate CDI, metronidazole is the
drug of choice. Vancomycin should be preferred for an initial episode
of severe CDI. In case of existence of ileus, megacolon, hypotension or
shock, vancomycin at higher doses (500 mg 4 times per day) plus
metronidazole can be administered. In recurrent CDI, the
recommendations for first recurrence are the same as for initial
episode. However, in second recurrence, vancomycin (in a tapered and/or
pulsed regimen) is the drug of choice.[132]
The
data on alternative treatment options for CDI are limited in
haematology patients and HSCT recipients. With the use of fidaxomicin,
clinical response and recurrence rates were found to be comparable to
that of conventional therapy.[133]
However, fidaxomicin was associated with a lower recurrence rate of CDI
associated with NAP1 strains.[134]
In a recent post hoc analysis, fidaxomicin was found to be superior to
vancomycin for treatment of CDI in patients with cancer in terms of
shorter time to resolution of diarrhoea, higher cure and sustained
response rates, and fewer recurrences.[135]
Screening of MDR Bacteria in Patients
with HSCT
Infection
prevention
and control measures such as hand hygiene, contact barrier precautions,
isolation, and appropriate environmental cleaning are crucial to deal
with the spread of MDR bacteria in haematology settings.[144-146]
Active surveillance can help to identify individuals colonised with MDR
pathogens. However, it is not clearly defined whether an
active-surveillance for MDR bacteria as an additional strategy to
infection control procedures is beneficial to prevent
health-care-associated
transmission.[144,147-149]
Colonisation may persist for months in the case of severe underlying
disorders, prolonged or recurrent antimicrobial exposure, and presence
of invasive devices.[144,150]
Patient populations for targeted screening, as well as ideal screening
method and timing of surveillance, are not definitely determined, but
can be chosen among those considered to have risk factors for
colonization with MDR pathogens, such as prolonged hospital stay,
exposure to antimicrobials, ICU stay or transfer from settings known to
have high MDR bacteria rates. Another approach is to obtain
surveillance cultures from each patient admitted to the settings with
high prevalence of MDR pathogens. While some centres establish weekly
surveillance cultures, others choose to obtain cultures at the time of
admission and/or whenever risk factors emerge for colonisation of MDR
bacteria.[144,150-153]
Screening
for MRSA colonisation is not routinely performed, but can be
established if MRSA rates remain to be high despite effective
implementation of infection control measures. In such circumstances,
MRSA surveillance cultures should be obtained on admission and
thereafter (e.g. weekly) with or without concomitant decolonization.[144,154-156]
VRE surveillance cultures can be considered in case of ongoing spread
of VRE in an HSCT unit to identify colonised patients.[144,152]
Active
surveillance cultures for MDR-GNBs can be used in units with high rates
of MDR-GNB infections. A point prevalence survey is recommended if
previously unnoticed cases with CRE are identified by the review of
microbiology reports for the preceding 6-12 months.[144,146] In a retrospective nationwide
survey from Italy, documented carbapenem-resistant K. pneumoniae
(CRKp) colonization before or after HSCT was determined to be followed
by infection in 25.8% of autologous HSCT and 39.2% of allogeneic HSCT
recipients; and infection-related mortality rates were stated as 16% in
autologous HSCT and 64.4% in allogeneic HSCT patients.[44]
In endemic settings, screening for CRKp before transplantation prior to
hospital admission and weekly after transplantation for those who
remain negative in case of isolation of CRKp in that unit is
recommended.[157,158] Recent
reports have revealed
that decolonization with aminoglycosides or colistin could succeed in
patients colonised with CRE.[157,159-163]
Nevertheless, development of resistance to these agents is of concern,
and patients can be recolonized after gastrointestinal decolonization.[158,159,164-166]
Management of Febrile Neutropenia in
the Era of Resistant Bacterial Infections
For
empirical
antibacterial treatment in febrile neutropenia, escalation or
de-escalation approach can be used. In escalation strategy, initial
therapy targets activity against Enterobacteriaceae and P. aeruginosa,
but, ESBL- and carbapenemase-producing gram-negative bacilli and
drug-resistant non-fermentative bacteria remain out of empirical
coverage. In case of development of clinical deterioration or isolation
of a resistant pathogen from clinical samples, the spectrum of
antibacterial coverage must be broadened. In de-escalation strategy,
initial regimen targets to cover drug-resistant pathogens, and once the
microbiological data become available, therapy is de-escalated to an
appropriate narrower spectrum. Escalation strategy may be considered
for patients followed in a centre where MDR pathogens are rarely seen
at the onset of febrile neutropenia and for those without any specific
risk factors for resistant bacterial infections. De-escalation strategy
may be used for febrile neutropenic patients having risk factors for
resistant bacterial infections, such as previous infection or known
colonisation with ESBL-producing gram-negative bacteria, residents of a
centre where MDR pathogens are common, and also for those presenting
with septic shock and pneumonia. Initial regimen in de-escalation
strategy may include monotherapy with a carbapenem or combination
therapy with an anti-pseudomonal beta-lactam agent and an
aminoglycoside/quinolone or combination therapy with colistin and a
beta-lactam agent/rifampicin. If risk factors for resistant
gram-positive infections are present, early coverage with a
glycopeptide or newer agents (linezolid, daptomycin, tigecycline) with
activity against glycopeptide non-susceptible gram-positive pathogens
should be considered. Patients with suspicion of catheter-related
infection, known colonisation with MRSA, VRE, and PRSP, hemodynamic
instability, severe sepsis, septic shock, presence of skin and soft
tissue infection and pneumonia are accepted as candidates for
additional antibiotics against resistant gram-positive pathogens.[12,27-29,51]
Conclusions
The
emergence of
infections with resistant bacterial pathogens is associated with trends
towards poor outcomes, prolonged hospital stay, more frequent ICU
admissions, and increased treatment costs in haematology patients.[26,45,136-138]
Moreover, the bacterial resistance complicates the use of standard
antimicrobial regimens in febrile HSCT recipients. Antimicrobial
treatment approach for neutropenic or chronically immunosuppressed HSCT
recipients with GVHD necessitates careful evaluation of patients;
detailed knowledge on local epidemiological data on antibacterial
resistance; close monitoring of the emergence of resistance in
bacterial pathogens; and use of robust treatment options in the context
of a rational antimicrobial stewardship program.[9,139,140]
Convenient infection control measures and appropriate vaccination
schedules should be implemented to prevent patients from exposure to
pathogens.[9,30,50,141-143]
Besides, effective attempts should be provided in the development of
new antibacterial agents and immune augmentation strategies to cope
with resistant bacterial pathogens.[9]
References
- Giralt S. Allogeneic
hematopoietic progenitor cell
transplantation for the treatment of chronic myelogenous leukemia in
the era of tyrosine kinase inhibitors: lessons learned to date. Clin
Lymphoma Myeloma 2007; 7 Suppl 3: S102-4. https://doi.org/10.3816/CLM.2007.s.009
PMid:17382018

- Dreger
P, Corradini P, Kimby E, et al. Chronic Leukemia Working Party of the
EBMT. Indications for allogeneic stem cell transplantation in chronic
lymphocytic leukemia: the EBMT transplant consensus. Leukemia 2007; 21:
12-7. https://doi.org/10.1038/sj.leu.2404441
PMid:17109028

- Davies
JK, Guinan EC. An update on the management of severe idiopathic
aplastic anaemia in children. Br J Haematol 2007; 136: 549-64. https://doi.org/10.1111/j.1365-2141.2006.06461.x
PMid:17214739

- Mackall
C, Fry T, Gress R, Peggs K, Storek J, Toubert A. Background to
hematopoietic cell transplantation, including post transplant immune
recovery. Bone Marrow Transplant 2009; 44: 457-62. https://doi.org/10.1038/bmt.2009.255
PMid:19861978

- Meijer
E, Dekker AW, Lokhorst HM, Petersen EJ, Nieuwenhuis HK, Verdonck LF.
Low incidence of infectious complications after nonmyeloablative
compared with myeloablative allogeneic stem cell transplantation.
Transpl Infect Dis 2004; 6: 171-8. https://doi.org/10.1111/j.1399-3062.2004.00075.x
PMid:15762935

- Junghanss
C, Boeckh M, Carter RA, Sandmaier BM, Maris MB, Maloney DG, Chauncey T,
McSweeney PA, Little MT, Corey L, Storb R. Incidence and outcome of
cytomegalovirus infections following nonmyeloablative compared with
myeloablative allogeneic stem cell transplantation, a matched control
study. Blood 2002; 99: 1978-85. https://doi.org/10.1182/blood.V99.6.1978
PMid:11877269

- Junghanss
C, Marr KA, Carter RA, Sandmaier BM, Maris MB, Maloney DG, Chauncey T,
McSweeney PA, Storb R. Incidence and outcome of bacterial and fungal
infections following nonmyeloablative compared with myeloablative
allogeneic hematopoietic stem cell transplantation: a matched control
study. Biol Blood Marrow Transplant 2002; 8: 512-20. https://doi.org/10.1053/bbmt.2002.v8.pm12374456
PMid:12374456

- Baron
F, Sandmaier BM. Chimerism and outcomes after allogeneic hematopoietic
cell transplantation following nonmyeloablative conditioning. Leukemia
2006; 20: 1690-700. https://doi.org/10.1038/sj.leu.2404335
PMid:16871276

- Kontoyiannis
DP, Lewis RE, Marr K. The burden of bacterial and viral infections in
hematopoietic stem cell transplant. Biol Blood Marrow Transplant 2009;
15(1 Suppl): 128-133. https://doi.org/10.1016/j.bbmt.2008.10.005
PMid:19147091

- Dettenkofer
M, Ebner W, Bertz H, Babikir R, Finke J, Frank U, et al. Surveillance
of nosocomial infections in adult recipients of allogeneic and
autologous bone marrow and peripheral blood stem-cell transplantation.
Bone Marrow Transplant 2003; 31: 795-801. https://doi.org/10.1038/sj.bmt.1703920
PMid:12732887

- Engelhart
S, Glasmacher A, Exner M, Kramer MH. Surveillance for nosocomial
infections and fever of unknown origin among adult hematology-oncology
patients. Infect Control Hosp Epidemiol 2002; 23: 244-8. https://doi.org/10.1086/502043
PMid:12026148

- Alp S, Akova M.
Management of febrile neutropenia in the era of bacterial resistance.
Ther Adv Infect Dis 2013; 1: 37-43. https://doi.org/10.1177/2049936113475610 PMid:25165543
PMCid:PMC4040719

- Harbarth
S, Harris AD, Carmeli Y, Samore MH. Parallel analysis of individual and
aggregated data on antibiotic exposure and resistance in gram-negative
bacilli. Clin Infect Dis 2001; 33: 1462-1468. https://doi.org/10.1086/322677
PMid:11588690

- Mebis
J, Goossens H, Berneman ZN. Antibiotic management of febrile
neutropenia: current developments and future directions. J Chemother
2010; 22: 5-12. https://doi.org/10.1179/joc.2010.22.1.5
PMid:20227985

- Bousquet
A, Malfuson JV, Sanmartin N, Konopacki J, MacNab C, Souleau B, de Revel
T, Elouennass M, Samson T, Soler C, Foissaud V, Martinaud C. An 8-year
survey of strains identified in blood cultures in a clinical
haematology unit. Clin Microbiol Infect 2014; 20: O7-12. https://doi.org/10.1111/1469-0691.12294
PMid:23826912

- Garnica
M, Nouér SA, Pellegrino FL, Moreira BM, Maiolino A, Nucci M.
Ciprofloxacin prophylaxis in high risk neutropenic patients: effects on
outcomes, antimicrobial therapy and resistance. BMC Infect Dis 2013;
13: 356. https://doi.org/10.1186/1471-2334-13-356 PMid:23899356
PMCid:PMC3729823

- Therriault
BL, Wilson JW, Barreto JN, Estes LL. Characterization of bacterial
infections in allogeneic hematopoietic stem cell transplant recipients
who received prophylactic levofloxacin with either penicillin or
doxycycline. Mayo Clin Proc 2010; 85: 711-8. https://doi.org/10.4065/mcp.2010.0006
PMid:20675508 PMCid:PMC2912731

- Schelenz
S, Nwaka D, Hunter PR. Longitudinal surveillance of bacteraemia in
haematology and oncology patients at a UK cancer centre and the impact
of ciprofloxacin use on antimicrobial resistance. J Antimicrob
Chemother 2013; 68: 1431-8. https://doi.org/10.1093/jac/dkt002
PMid:23396855

- Kern
WV, Steib-Bauert M, de With K, Reuter S, Bertz H, Frank U, von Baum H.
Fluoroquinolone consumption and resistance in haematology-oncology
patients: ecological analysis in two university hospitals 1999-2002. J
Antimicrob Chemother 2005; 55: 57-60. https://doi.org/10.1093/jac/dkh510
PMid:15574472

- Castagnola
E, Haupt R, Micozzi A, Caviglia I, Testi AM, Giona F, Parodi S,
Girmenia C. Differences in the proportions of fluoroquinolone-resistant
gram-negative bacteria isolated from bacteraemic children with cancer
in two Italian centres. Clin Microbiol Infect 2005; 11: 505-7. https://doi.org/10.1111/j.1469-0691.2005.01114.x
PMid:15882204

- Rangaraj
G, Granwehr BP, Jiang Y, Hachem R, Raad I. Perils of quinolone exposure
in cancer patients: breakthrough bacteremia with multidrug-resistant
organisms. Cancer 2010; 116: 967-73. https://doi.org/10.1002/cncr.24812
PMid:20052728

- MacDougall
C, Powell JP, Johnson CK, Edmond MB, Polk RE. Hospital and community
fluoroquinolone use and resistance in Staphylococcus aureus and
Escherichia coli in 17 US hospitals. Clin Infect Dis 2005; 41: 435-440.
https://doi.org/10.1086/432056
PMid:16028149

- Muto
CA, Pokrywka M, Shutt K, Mendelsohn AB, Nouri K, Posey K, et al. A
large outbreak of Clostridium difficile-associated disease with an
unexpected proportion of deaths and colectomies at a teaching hospital
following increased fluoroquinolone use. Infect Control Hosp Epidemiol
2005; 26: 273-280. https://doi.org/10.1086/502539
PMid:15796280

- Park
SY, Kang CI, Joo EJ, Ha YE, Wi YM, Chung DR, et al. Risk factors for
multidrug resistance in nosocomial bacteremia caused by
extended-spectrum ß-lactamase-producing Escherichia coli and Klebsiella
pneumoniae. Microb Drug Resist 2012; 18: 518-524. https://doi.org/10.1089/mdr.2012.0067
PMid:22742454

- Pépin
J, Saheb N, Coulombe MA, Alary ME, Corriveau MP, Authier S, et al.
Emergence of fluoroquinolones as the predominant risk factor for
Clostridium difficile-associated diarrhea: a cohort study during an
epidemic in Quebec. Clin Infect Dis 2005; 41: 1254-1260. https://doi.org/10.1086/496986
PMid:16206099

- Trubiano
JA, Worth LJ, Thursky KA, Slavin MA. The prevention and management of
infections due to multidrug resistant organisms in haematology
patients. Br J Clin Pharmacol 2015; 79: 195-207. https://doi.org/10.1111/bcp.12310
PMid:24341410 PMCid:PMC4309626

- Averbuch
D, Orasch C, Cordonnier C, Livermore DM, Mikulska M, Viscoli C, Gyssens
IC, Kern WV, Klyasova G, Marchetti O, Engelhard D, Akova M; ECIL4, a
joint venture of EBMT, EORTC, ICHS, ESGICH/ESCMID and ELN. European
guidelines for empirical antibacterial therapy for febrile neutropenic
patients in the era of growing resistance: summary of the 2011 4th
European Conference on Infections in Leukemia. Haematologica 2013; 98:
1826-35. https://doi.org/10.3324/haematol.2013.091025
PMid:24323983 PMCid:PMC3856957

- Averbuch
D, Cordonnier C, Livermore DM, Mikulska M, Orasch C, Viscoli C, Gyssens
IC, Kern WV, Klyasova G, Marchetti O, Engelhard D, Akova M; ECIL4, a
joint venture of EBMT, EORTC, ICHS, ESGICH/ESCMID and ELN. Targeted
therapy against multi-resistant bacteria in leukemic and hematopoietic
stem cell transplant recipients: guideliens of the 4th European
Conference on Infections in Leukemia (ECIL-4, 2011). Haematologica
2013; 98: 1836-47. https://doi.org/10.3324/haematol.2013.091330
PMid:24323984 PMCid:PMC3856958

- Mikulska
M, Del Bono V, Viscoli C. Bacterial infections in hematopoietic stem
cell transplantation recipients. Curr Opin Hematol 2014; 21: 451-8. https://doi.org/10.1097/MOH.0000000000000088
PMid:25295742

- Ruhnke
M, Arnold R, Gastmeier P. Infection control issues in patients with
haematological malignancies in the era of multidrug-resistant bacteria.
Lancet Oncol 2014; 15: e606-19. https://doi.org/10.1016/S1470-2045(14)70344-4

- Gustinetti
G, Mikulska M. Bloodstream infections in neutropenic cancer patients: A
practical update. Virulence 2016; 7: 280-97.
https://doi.org/10.1080/21505594.2016.1156821
PMid:27002635

- Freifeld
AG, Bow EJ, Sepkowitz KA, Boeckh MJ, Ito JI, Mullen CA, Raad II,
Rolston KV, Young JA, Wingard JR, Infectious Diseases Society of
America. Clinical practice guideline for the use of antimicrobial
agents in neutropenic patients with cancer: 2010 Update by the
Infectious Diseases Society of America. Clin Infect Dis 2011; 52:
427-31. https://doi.org/10.1093/cid/ciq147
PMid:21205990

- Ramphal R. Changes in
the etiology of bacteremia in febrile neutropenic patients and the
susceptibilities of the currently isolated pathogens. Clin Infect Dis
2004; 39(Suppl 1): S25-31. https://doi.org/10.1086/383048
PMid:15250017

- Zinner SH. Changing
epidemiology of infections in patients with neutropenia and cancer:
emphasis on gram-positive and resistant bacteria. Clin Infect Dis 1999;
29: 490-494. https://doi.org/10.1086/598620
PMid:10530434

- Wisplinghoff H, Seifert
H, Wenzel RP, Edmond MB. Current trends in the epidemiology of
nosocomial bloodstream infections in patients with hematological
malignancies and solid neoplasms in hospitals in the United States.
Clin Infect Dis 2003; 36: 1103-1110. https://doi.org/10.1086/374339
PMid:12715303

- Aubron C, Poirel L,
Fortineau N, Nicolas P, Collet L, Nordmann P. Nosocomial spread of
Pseudomonas aeruginosa isolates expressing the metallo-beta-lactamase
VIM-2 in a hematology unit of a French hospital. Microb Drug Resist
2005; 11: 254-259. https://doi.org/10.1089/mdr.2005.11.254
PMid:16201928

- Cattaneo C, Quaresmini
G, Casari S, Capucci MA, Micheletti M, Borlenghi E, et al. Recent
changes in bacterial epidemiology and the emergence of
fluoroquinolone-resistant Escherichia coli among patients with
haematological malignancies: results of a prospective study on 823
patients at a single institution. J Antimicrob Chemother 2008; 61:
721-728. https://doi.org/10.1093/jac/dkm514
PMid:18218645

- Chen CY, Tang JL, Hsueh
PR, Yao M, Huang SY, Chen YC, et al. Trends and antimicrobial
resistance of pathogens causing bloodstream infections among febrile
neutropenic adults with hematological malignancy. J Formos Med Assoc
2004; 103: 526-532. PMid:15318274

- Gaynes
R, Edwards JR. Overview of nosocomial infections caused by
gram-negative bacilli. Clin Infect Dis 2005; 41: 848-854.
https://doi.org/10.1086/432803
PMid:16107985

- Gudiol
C, Tubau F, Calatayud L, Garcia-Vidal C, Cisnal M, Sánchez-Ortega I, et
al. Bacteraemia due to multidrug-resistant gram-negative bacilli in
cancer patients: risk factors, antibiotic therapy and outcomes. J
Antimicrob Chemother 2011; 66: 657-63.
https://doi.org/10.1093/jac/dkq494
PMid:21193475

- Gudiol
C, Bodro M, Simonetti A, Tubau F, González-Barca E, Cisnal M,
Domingo-Domenech E, Jiménez L, Carratalà J. Changing aetiology,
clinical features, antimicrobial resistance, and outcomes of
bloodstream infection in neutropenic cancer patients. Clin Microbiol
Infect 2013; 19: 474-9.
https://doi.org/10.1111/j.1469-0691.2012.03879.x
PMid:22524597

- Hakki M, Limaye AP, Kim
HW, Kirby KA, Corey L, Boeckh M. Invasive Pseudomonas aeruginosa
infections: high rate of recurrence and mortality after hematopoietic
cell transplantation. Bone Marrow Transplant 2007; 39: 687-693.
https://doi.org/10.1038/sj.bmt.1705653
PMid:17401395

- Oliveira
AL, de Souza M, Carvalho-Dias VM, Ruiz MA, Silla L, Tanaka PY, et al.
Epidemiology of bacteremia and factors associated with
multi-drug-resistant gram-negative bacteremia in hematopoietic stem
cell transplant recipients. Bone Marrow Transplant 2007; 39: 775-781.
https://doi.org/10.1038/sj.bmt.1705677
PMid:17438585

- Girmenia
C, Rossolini GM, Piciocchi A, Bertaina A, Pisapia G, Pastore D, Sica S,
Severino A, Cudillo L, Ciceri F, Scimè R, Lombardini L, Viscoli C,
Rambaldi A; Gruppo Italiano Trapianto Midollo Osseo (GITMO); Gruppo
Italiano Trapianto Midollo Osseo GITMO. Infections by
carbapenem-resistant Klebsiella pneumoniae in SCT recipients: a
nationwide retrospective survey from Italy. Bone Marrow
Transplant.2015; 50: 282-8. https://doi.org/10.1038/bmt.2014.231
PMid:25310302

- Gudiol C, Calatayud L,
Garcia-Vidal C, Lora-Tamayo J, Cisnal M, Duarte R, Arnan M, Marin M,
Carratalà J, Gudiol F. Bacteraemia due to extended-spectrum
beta-lactamase-producing Escherichia coli (ESBL-EC) in cancer patients:
clinical features, risk factors, molecular epidemiology and outcome. J
Antimicrob Chemother 2010; 65: 333-41.
https://doi.org/10.1093/jac/dkp411
PMid:19959544

- Morris
PG, Hassan T, McNamara M, Hassan A, Wiig R, Grogan L, et al. Emergence
of MRSA in positive blood cultures from patients with febrile
neutropenia-a cause for concern. Support Care Cancer 2008;
16:1085-1088. https://doi.org/10.1007/s00520-007-0398-5
PMid:18274787

- Weinstock DM, Conlon M,
Iovino C, Aubrey T, Gudiol C, Riedel E, et al. Colonization,
bloodstream infection, and mortality caused by vancomycin-resistant
enterococcus early after allogeneic hematopoietic stem cell transplant.
Biol Blood Marrow Transplant 2007; 13: 615-621.
https://doi.org/10.1016/j.bbmt.2007.01.078
PMid:17448922

- Carratala J, Roson B,
Fernandez-Sevilla A, Alcaide F, Gudiol F. Bacteremic pneumonia in
neutropenic patients with cancer: causes, empirical antibiotic therapy,
and outcome. Arch Intern Med 1998; 158: 868-872.
https://doi.org/10.1001/archinte.158.8.868
PMid:9570172

- Mikulska M, Viscoli C,
Orasch C, Livermore DM, Averbuch D, Cordonnier C, Akova M; Fourth
European Conference on Infections in Leukemia Group (ECIL-4), a joint
venture of EBMT, EORTC, ICHS, ELN and ESGICH/ESCMID. Aetiology and
resistance in bacteraemias among adult and paediatric haematology and
cancer patients. J Infect 2014; 68: 321-31.
https://doi.org/10.1016/j.jinf.2013.12.006
PMid:24370562

- Tatarelli P, Mikulska
M. Multidrug-resistant bacteria in hematology patients: emerging
threats. Future Microbiol 2016; 11: 767-80.
https://doi.org/10.2217/fmb-2015-0014
PMid:27196948

- Balletto
E, Mikulska M. Bacterial Infections in hematopoietic stem cell
transplant recipients. Mediterr J Hematol Infect Dis 2015; 7: e2015045.
https://doi.org/10.4084/mjhid.2015.045
PMid:26185610 PMCid:PMC4500472

- Akova
M. Epidemiology of antimicrobial resistance in bloodstream infections.
Virulence 2016; 7: 252-66.
https://doi.org/10.1080/21505594.2016.1159366
PMid:26984779

- Kara
O, Zarakolu P, Ascioglu S, Etgul S, Uz B, Buyukasik Y, Akova M.
Epidemiology and emerging resistance in bacterial bloodstream
infections in patients with hematologic malignancies. Infect Dis (Lond)
2015; 47: 686-93. https://doi.org/10.3109/23744235.2015.1051105
PMid:26024284

- Paterson DL. Resistance
in gram-negative
bacteria: Enterobacteriaceae. Am J Infect Control 2006; 34(5 Suppl 1):
S20-8; discussion S64-73.

- Livermore DM. Current
epidemiology and growing resistance of gram-negative pathogens. Korean
J Intern Med 2012; 27: 128-42.
https://doi.org/10.3904/kjim.2012.27.2.128
PMid:22707882
PMCid:PMC3372794

- Livermore DM, Hope R,
Mushtaq S, Warner
M. Orthodox and unorthodox clavulanate combinations against
extended-spectrum beta-lactamase producers. Clin Microbiol Infect 2008;
14 Suppl 1: 189-93. https://doi.org/10.1111/j.1469-0691.2007.01858.x
PMid:18154546

- Guh AY, Bulens SN, Mu
Y, Jacob JT, Reno J, Scott J, Wilson LE, Vaeth E, Lynfield R, Shaw KM,
Vagnone PM, Bamberg WM, Janelle SJ, Dumyati G, Concannon C, Beldavs Z,
Cunningham M, Cassidy PM, Phipps EC, Kenslow N, Travis T, Lonsway D,
Rasheed JK, Limbago BM, Kallen AJ. Epidemiology of carbapenem-resistant
Enterobacteriaceae in 7 US communities, 2012-2013. JAMA 2015; 314:
1479-87. https://doi.org/10.1001/jama.2015.12480
PMid:26436831

- Nordmann P, Poirel L.
The difficult-to-control spread of carbapenemase producers among
Enterobacteriaceae worldwide. Clin Microbiol Infect 2014; 20: 821-30.
https://doi.org/10.1111/1469-0691.12719
PMid:24930781

- Cornaglia G,
Giamarellou H, Rossolini GM.
Metallo-ß-lactamases: a last frontier for ß-lactams? Lancet Infect Dis
2011; 11: 381-93. https://doi.org/10.1016/S1473-3099(11)70056-1

- Liu
YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B,
Huang X, Yu LF, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu
JH, Shen J. Emergence of plasmid-mediated colistin resistance mechanism
MCR-1 in animals and human beings in China: a microbiological and
molecular biological study. Lancet Infect Dis 2016; 16):
161-8.

- Hasman
H, Hammerum AM, Hansen F, Hendriksen RS, Olesen B, Agersø Y, Zankari E,
Leekitcharoenphon P, Stegger M, Kaas RS, Cavaco LM, Hansen DS,
Aarestrup FM, Skov RL. Detection of mcr-1 encoding plasmid-mediated
colistin-resistant Escherichia coli isolates from human bloodstream
infection and imported chicken meat, Denmark 2015. Euro Surveill 2015;
20(49). https://doi.org/10.2807/1560-7917.ES.2015.20.49.30085
PMid:26676364

- Malhotra-Kumar S,
Xavier BB, Das AJ, Lammens C, Hoang HT, Pham NT, Goossens H.
Colistin-resistant Escherichia coli harbouring mcr-1 isolated from food
animals in Hanoi, Vietnam. Lancet Infect Dis. 2016 Mar;16(3):286-7.
https://doi.org/10.1016/S1473-3099(16)00014-1

- Malhotra-Kumar
S, Xavier BB, Das AJ, Lammens C, Butaye P, Goossens H. Colistin
resistance gene mcr-1 harboured on a multidrug resistant plasmid.
Lancet Infect Dis 2016; 16: 283-4.
https://doi.org/10.1016/S1473-3099(16)00012-8

- Falgenhauer
L, Waezsada SE, Yao Y, Imirzalioglu C, Käsbohrer A, Roesler U, Michael
GB, Schwarz S, Werner G, Kreienbrock L, Chakraborty T; RESET
consortium. Colistin resistance gene mcr-1 in extended-spectrum
ß-lactamase-producing and carbapenemase-producing gram-negative
bacteria in Germany. Lancet Infect Dis 2016; 16: 282-3.
https://doi.org/10.1016/S1473-3099(16)00009-8

- Perrin-Guyomard
A, Bruneau M, Houée P, Deleurme K, Legrandois P, Poirier C, Soumet C,
Sanders P. Prevalence of mcr-1 in commensal Escherichia coli from
French livestock, 2007 to 2014. Euro Surveill 2016; 21(6).
https://doi.org/10.2807/1560-7917.ES.2016.21.6.30135
PMid:26898350

- Paterson DL, Harris PN.
Colistin resistance: a major breach in our last line of defence. Lancet
Infect Dis 2016; 16: 132-3.
https://doi.org/10.1016/S1473-3099(15)00463-6

- Tse H,
Yuen KY. Dissemination of the mcr-1 colistin resistance gene. Lancet
Infect Dis 2016; 16: 145-6.
https://doi.org/10.1016/S1473-3099(15)00532-0

- Webb HE,
Granier SA, Marault M, Millemann Y, den Bakker HC, Nightingale KK,
Bugarel M, Ison SA, Scott HM, Loneragan GH. Dissemination of the mcr-1
colistin resistance gene. Lancet Infect Dis 2016; 16: 144-5.
https://doi.org/10.1016/S1473-3099(15)00538-1

- Du H,
Chen L, Tang YW, Kreiswirth BN. Emergence of the mcr-1 colistin
resistance gene in carbapenem-resistant Enterobacteriaceae. Lancet
Infect Dis 2016; 16: 287-8.
https://doi.org/10.1016/S1473-3099(16)00056-6

- Stoesser
N, Mathers AJ, Moore CE, Day NP, Crook DW. Colistin resistance gene
mcr-1 and pHNSHP45 plasmid in human isolates of Escherichia coli and
Klebsiella pneumoniae. Lancet Infect Dis 2016; 16: 285-6.
https://doi.org/10.1016/S1473-3099(16)00010-4

- Cantón R,
Akóva M, Carmeli Y, Giske CG, Glupczynski Y, Gniadkowski M, Livermore
DM, Miriagou V, Naas T, Rossolini GM, Samuelsen Ø, Seifert H, Woodford
N, Nordmann P; European Network on Carbapenemases. Rapid evolution and
spread of carbapenemases among Enterobacteriaceae in Europe. Clin
Microbiol Infect 2012; 18: 413-31.
https://doi.org/10.1111/j.1469-0691.2012.03821.x
PMid:22507109

- Hu L, Zhong Q, Shang Y,
Wang H, Ning C, Li Y, Hang Y, Xiong J, Wang X, Xu Y, Qin Z, Parsons C,
Wang L, Yu F. The prevalence of carbapenemase genes and
plasmid-mediated quinolone resistance determinants in
carbapenem-resistant Enterobacteriaceae from five teaching hospitals in
central China. Epidemiol Infect 2014; 142: 1972-7.
https://doi.org/10.1017/S0950268813002975
PMid:24252194

- Munoz-Price LS, Poirel
L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, Cornaglia G, Garau J,
Gniadkowski M, Hayden MK, Kumarasamy K, Livermore DM, Maya JJ, Nordmann
P, Patel JB, Paterson DL, Pitout J, Villegas MV, Wang H, Woodford N,
Quinn JP. Clinical epidemiology of the global expansion of Klebsiella
pneumoniae carbapenemases. Lancet Infect Dis 2013; 13: 785-96.
https://doi.org/10.1016/S1473-3099(13)70190-7

- Tseng IL,
Liu YM, Wang SJ, Yeh HY, Hsieh CL, Lu HL, Tseng YC, Mu JJ. Emergence of
carbapenemase producing Klebsiella pneumonia and spread of KPC-2 and
KPC-17 in Taiwan: A nationwide study from 2011 to 2013. PLoS One 2015;
10: e0138471. https://doi.org/10.1371/journal.pone.0138471
PMid:26384242 PMCid:PMC4575059

- Monaco M, Giani T,
Raffone
M, Arena F, Garcia-Fernandez A, Pollini S; Network EuSCAPE-Italy,
Grundmann H, Pantosti A, Rossolini GM. Colistin resistance superimposed
to endemic carbapenem-resistant Klebsiella pneumoniae: a rapidly
evolving problem in Italy, November 2013 to April 2014. Euro Surveill
2014; 19(42). https://doi.org/10.2807/1560-7917.ES2014.19.42.20939
PMid:25358041

- Giacobbe DR, Del Bono
V, Trecarichi EM, De Rosa FG, Giannella M, Bassetti M, Bartoloni A,
Losito AR, Corcione S, Bartoletti M, Mantengoli E, Saffioti C, Pagani
N, Tedeschi S, Spanu T, Rossolini GM, Marchese A, Ambretti S, Cauda R,
Viale P, Viscoli C, Tumbarello M; ISGRI-SITA (Italian Study Group on
Resistant Infections of the Società Italiana Terapia Antinfettiva).
Risk factors for bloodstream infections due to colistin-resistant
KPC-producing Klebsiella pneumoniae: results from a multicenter
case-control-control study. Clin Microbiol Infect 2015; 21: 1106.e1-8.
https://doi.org/10.1016/j.cmi.2015.08.001 PMid:26278669

- European Centre for
Disease Prevention and Control (ECDC). Annual epidemiological report
2014. Antimicrobial resistance and healthcare-associated infections.
2015. Available at:
http://ecdc.europa.eu/en/publications/Publications/antimicrobial-resistance-annual-epidemiological-report.pdf.
- Potron
A, Poirel L, Nordmann P. Emerging broad-spectrum resistance in
Pseudomonas aeruginosa and Acinetobacter baumannii: Mechanisms and
epidemiology. Int J Antimicrob Agents 2015; 45: 568-85.
https://doi.org/10.1016/j.ijantimicag.2015.03.001
PMid:25857949

- Jean
SS, Lee WS, Yu KW, Liao CH, Hsu CW, Chang FY, Ko WC, Chen RJ, Wu JJ,
Chen YH, Chen YS, Liu JW, Lu MC, Lam C, Liu CY, Hsueh PR. Rates of
susceptibility of carbapenems, ceftobiprole, and colistin against
clinically important bacteria collected from intensive care units in
2007: Results from the Surveillance of Multicenter Antimicrobial
Resistance in Taiwan (SMART). J Microbiol Immunol Infect 2015 Jan 10.
pii: S1684-1182(15)00021-3.

- Dortet L, Poirel L,
Nordmann
P. Worldwide dissemination of the NDM-type carbapenemases in
gram-negative bacteria. Biomed Res Int 2014; 2014: 249856.
https://doi.org/10.1155/2014/249856
PMid:24790993 PMCid:PMC3984790

- Durakovic
N, Radojcic V, Boban A, Mrsic M, Sertic D, Serventi-Seiwerth R, Nemet
D, Labar B. Efficacy and safety of colistin in the treatment of
infections caused by multidrug-resistant Pseudomonas aeruginosa in
patients with hematologic malignancy: a matched pair analysis. Intern
Med 2011; 50: 1009-13. https://doi.org/10.2169/internalmedicine.50.4270
PMid:21532223

- Micol JB, de Botton S,
Guieze R, Coiteux
V, Darre S, Dessein R, Leroy O, Yakoub-Agha I, Quesnel B, Bauters F,
Beaucaire G, Alfandari S. An 18-case outbreak of drug-resistant
Pseudomonas aeruginosa bacteriemia in hematology patients.
Haematologica 2006; 91: 1134-8. PMid:16885056

- Kumar
A, Zarychanski R, Light B, Parrillo J, Maki D, Simon D, Laporta D,
Lapinsky S, Ellis P, Mirzanejad Y, Martinka G, Keenan S, Wood G, Arabi
Y, Feinstein D, Kumar A, Dodek P, Kravetsky L, Doucette S; Cooperative
Antimicrobial Therapy of Septic Shock (CATSS) Database Research Group.
Early combination antibiotic therapy yields improved survival compared
with monotherapy in septic shock: a propensity-matched analysis. Crit
Care Med 2010; 38: 1773-85.
https://doi.org/10.1097/CCM.0b013e3181eb3ccd
PMid:20639750

- Safdar
N, Handelsman J, Maki DG. Does combination antimicrobial therapy reduce
mortality in gram-negative bacteraemia? A meta-analysis. Lancet Infect
Dis 2004; 4: 519-27. https://doi.org/10.1016/S1473-3099(04)01108-9

- Martínez
JA, Cobos-Trigueros N, Soriano A, Almela M, Ortega M, Marco F, Pitart
C, Sterzik H, Lopez J, Mensa J. Influence of empiric therapy with a
beta-lactam alone or combined with an aminoglycoside on prognosis of
bacteremia due to gram-negative microorganisms. Antimicrob Agents
Chemother 2010; 54: 3590-6. https://doi.org/10.1128/AAC.00115-10
PMid:20585123 PMCid:PMC2934963

- Hogg GM, Barr JG, Webb
CH.
In-vitro activity of the combination of colistin and rifampicin against
multidrug-resistant strains of Acinetobacter baumannii. J Antimicrob
Chemother 1998; 41: 494-5. https://doi.org/10.1093/jac/41.4.494
PMid:9598783

- Petrosillo N, Chinello
P, Proietti MF, Cecchini L, Masala M, Franchi C, Venditti M, Esposito
S, Nicastri E. Combined colistin and rifampicin therapy for
carbapenem-resistant Acinetobacter baumannii infections: clinical
outcome and adverse events. Clin Microbiol Infect 2005; 11: 682-3.
https://doi.org/10.1111/j.1469-0691.2005.01198.x
PMid:16008625

- Durante-Mangoni E,
Signoriello G, Andini R, Mattei A, De Cristoforo M, Murino P, Bassetti
M, Malacarne P, Petrosillo N, Galdieri N, Mocavero P, Corcione A,
Viscoli C, Zarrilli R, Gallo C, Utili R. Colistin and rifampicin
compared with colistin alone for the treatment of serious infections
due to extensively drug-resistant Acinetobacter baumannii: a
multicenter, randomized clinical trial. Clin Infect Dis 2013; 57:
349-58. https://doi.org/10.1093/cid/cit253
PMid:23616495

- Qureshi ZA, Paterson
DL, Potoski BA, Kilayko MC, Sandovsky G, Sordillo E, Polsky B,
Adams-Haduch JM, Doi Y. Treatment outcome of bacteremia due to
KPC-producing Klebsiella pneumoniae: superiority of combination
antimicrobial regimens. Antimicrob Agents Chemother 2012; 56: 2108-13.
https://doi.org/10.1128/AAC.06268-11
PMid:22252816 PMCid:PMC3318350

- Tenover
FC, Moellering RC Jr. The rationale for revising the Clinical and
Laboratory Standards Institute vancomycin minimal inhibitory
concentration interpretive criteria for Staphylococcus aureus. Clin
Infect Dis 2007; 44: 1208-1215. https://doi.org/10.1086/513203
PMid:17407040

- Chuang YY, Huang YC.
Molecular epidemiology of community-associated meticillin-resistant
Staphylococcus aureus in Asia. Lancet Infect Dis 2013; 13: 698-708.
https://doi.org/10.1016/S1473-3099(13)70136-1

- David MZ,
Daum RS. Community-associated methicillin-resistant Staphylococcus
aureus: epidemiology and clinical consequences of an emerging epidemic.
Clin Microbiol Rev 2010; 23: 616-87.
https://doi.org/10.1128/CMR.00081-09
PMid:20610826 PMCid:PMC2901661

- Laupland
KB, Lyytikäinen O, Søgaard M, Kennedy KJ, Knudsen JD, Ostergaard C,
Galbraith JC, Valiquette L, Jacobsson G, Collignon P, Schønheyder HC;
International Bacteremia Surveillance Collaborative. The changing
epidemiology of Staphylococcus aureus bloodstream infection: a
multinational population-based surveillance study. Clin Microbiol
Infect 2013; 19: 465-71.
https://doi.org/10.1111/j.1469-0691.2012.03903.x
PMid:22616816

- Skov RL, Jensen KS.
Community-associated meticillin-resistant Staphylococcus aureus as a
cause of hospital-acquired infections. J Hosp Infect 2009; 73: 364-70.
https://doi.org/10.1016/j.jhin.2009.07.004
PMid:19786313

- Rhee Y, Aroutcheva A,
Hota B, Weinstein RA, Popovich KJ. Evolving epidemiology of
Staphylococcus aureus bacteremia. Infect Control Hosp Epidemiol 2015;
36: 1417-22. https://doi.org/10.1017/ice.2015.213
PMid:26372679

- Stryjewski ME, Corey
GR. Methicillin-resistant Staphylococcus aureus: an evolving pathogen.
Clin Infect Dis. 2014 Jan;58 Suppl 1:S10-9. Hope R, Livermore DM, Brick
G, Lillie M, Reynolds R; BSAC Working Parties on Resistance
Surveillance. Non-susceptibility trends among staphylococci from
bacteraemias in the UK and Ireland, 2001-06. J Antimicrob Chemother
2008; 62 Suppl 2: ii65-74.

- Mermel LA, Allon M,
Bouza E,
Craven DE, Flynn P, O'Grady NP, Raad II, Rijnders BJ, Sherertz RJ,
Warren DK. Clinical practice guidelines for the diagnosis and
management of intravascular catheter-related infection: 2009 Update by
the Infectious Diseases Society of America. Clin Infect Dis 2009; 49:
1-45. https://doi.org/10.1086/599376
PMid:19489710 PMCid:PMC4039170

- Bennett
JE, Dolin R, Blaser MJ. Mandell, Douglas, and Bennett's Principle and
Practice of Infectious Diseases. 8th ed. Philadelphia (USA): Elsevier
Saunders; c2015. Chapter 201, Streptococcus pneumonia;
p.2310-27.
- EARS-Net:
European Centre for Disease Prevention and Control (ECDC),
Antimicrobial resistance interactive database (Internet). Stockholm
(Sweden): ECDC (cited 2015 Oct 22). Available from
http://ecdc.europa.eu/en/healthtopics/antimicrobial_resistance/database/Pages/database.aspx.
- Slipczuk
L, Codolosa JN, Davila CD, Romero-Corral A, Yun J, Pressman GS,
Figueredo VM. Infective endocarditis epidemiology over five decades: a
systematic review. PLoS One 2013; 8: e82665.
https://doi.org/10.1371/journal.pone.0082665
PMid:24349331
PMCid:PMC3857279

- Cordonnier C, Buzyn A,
Leverger G,
Herbrecht R, Hunault M, Leclercq R, Bastuji-Garin S; Club de Réflexion
sur les Infections en Onco-Hématologie. Epidemiology and risk factors
for gram-positive coccal infections in neutropenia: toward a more
targeted antibiotic strategy. Clin Infect Dis 2003; 36: 149-58.
https://doi.org/10.1086/345435
PMid:12522746

- Pfaller
MA, Jones RN, Marshall SA, Edmond MB, Wenzel RP. Nosocomial
streptococcal blood stream infections in the SCOPE Program: species
occurrence and antimicrobial resistance. The SCOPE Hospital Study
Group. Diagn Microbiol Infect Dis 1997; 29: 259-63.
https://doi.org/10.1016/S0732-8893(97)00159-4

- Pfaller
MA, Marshall SA, Jones RN. In vitro activity of cefepime and
ceftazidime against 197 nosocomial blood stream isolates of
streptococci: a multicenter sample. Diagn Microbiol Infect Dis 1997;
29: 273-6. https://doi.org/10.1016/S0732-8893(97)00139-9

- Sievert
DM, Ricks P, Edwards JR, Schneider A, Patel J, Srinivasan A, Kallen A,
Limbago B, Fridkin S; National Healthcare Safety Network (NHSN) Team
and Participating NHSN Facilities. Antimicrobial-resistant pathogens
associated with healthcare-associated infections: summary of data
reported to the National Healthcare Safety Network at the Centers for
Disease Control and Prevention, 2009-2010. Infect Control Hosp
Epidemiol 2013; 34: 1-14. https://doi.org/10.1086/668770
PMid:23221186

- Arias CA, Murray BE.
The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev
Microbiol 2012; 10: 266-78.
https://doi.org/10.1038/nrmicro2761 PMid:22421879
PMCid:PMC3621121

- O'Driscoll T, Crank
CW.
Vancomycin-resistant enterococcal infections: epidemiology, clinical
manifestations, and optimal management. Infect Drug Resist 2015; 8:
217-30. PMid:26244026 PMCid:PMC4521680

- Cattoir V,
Leclercq R. Twenty-five years of shared life with vancomycin-resistant
enterococci: is it time to divorce? J Antimicrob Chemother 2013; 68:
731-42. https://doi.org/10.1093/jac/dks469
PMid:23208830

- Micek ST. Alternatives
to vancomycin for the treatment of methicillin-resistant Staphylococcus
aureus infections. Clin Infect Dis 2007; 45(Suppl 3): S184-190.
https://doi.org/10.1086/519471
PMid:17712745

- Wahby
KA, Alangaden GJ. Daptomycin failure in a neutropenic leukemia patient
with Staphylococcus aureus meningitis. Leuk Lymphoma 2012; 53: 1610-2.
https://doi.org/10.3109/10428194.2012.661051
PMid:22390617

- Rolston
KV, Besece D, Lamp KC, Yoon M, McConnell SA, White P. Daptomycin use in
neutropenic patients with documented gram-positive infections. Support
Care Cancer 2014; 22: 7-14. https://doi.org/10.1007/s00520-013-1947-8
PMid:23975231

- Barber GR, Lauretta J,
Saez R. A febrile neutropenic patient with Enterococcus gallinarum
sepsis treated with daptomycin and gentamicin. Pharmacotherapy 2007;
27: 927-32. https://doi.org/10.1592/phco.27.6.927
PMid:17542774

- Yahav D, Lador A, Paul
M, Leibovici L. Efficacy and safety of tigecycline: a systematic review
and meta-analysis. J Antimicrob Chemother 2011; 66: 1963-71.
https://doi.org/10.1093/jac/dkr242
PMid:21685488

- Prasad
P, Sun J, Danner RL, Natanson C. Excess deaths associated with
tigecycline after approval based on noninferiority trials. Clin Infect
Dis 2012; 54: 1699-709. https://doi.org/10.1093/cid/cis270
PMid:22467668 PMCid:PMC3404716

- Alonso CD, Treadway
SB,
Hanna DB, Huff CA, Neofytos D, Carroll KC, Marr KA. Epidemiology and
outcomes of Clostridium difficile infections in hematopoietic stem cell
transplant recipients. Clin Infect Dis 2012; 54: 1053-63.
https://doi.org/10.1093/cid/cir1035
PMid:22412059 PMCid:PMC3309884

- Willems
L, Porcher R, Lafaurie M, Casin I, Robin M, Xhaard A, Andreoli AL,
Rodriguez-Otero P, Dhedin N, Socié G, Ribaud P, Peffault de Latour R.
Clostridium difficile infection after allogeneic hematopoietic stem
cell transplantation: incidence, risk factors, and outcome. Biol Blood
Marrow Transplant 2012; 18: 1295-301.
https://doi.org/10.1016/j.bbmt.2012.02.010
PMid:22387347

- Trifilio SM, Pi J,
Mehta J. Changing epidemiology of Clostridium difficile-associated
disease during stem cell transplantation. Biol Blood Marrow Transplant
2013; 19: 405-9. https://doi.org/10.1016/j.bbmt.2012.10.030
PMid:23219779

- Chakrabarti S, Lees A,
Jones SG, Milligan DW. Clostridium difficile infection in allogeneic
stem cell transplant recipients is associated with severe
graft-versus-host disease and non-relapse mortality. Bone Marrow
Transplant 2000; 26: 871-6. https://doi.org/10.1038/sj.bmt.1702627
PMid:11081387

- Bergogne-Bérézin E.
Treatment and prevention of antibiotic associated diarrhea. Int J
Antimicrob Agents 2000; 16: 521-6.
https://doi.org/10.1016/S0924-8579(00)00293-4

- Kyne L,
Hamel MB, Polavaram R, Kelly CP. Health care costs and mortality
associated with nosocomial diarrhea due to Clostridium difficile. Clin
Infect Dis 2002; 34: 346-53. https://doi.org/10.1086/338260
PMid:11774082

- McDonald LC, Killgore
GE, Thompson A, Owens RC Jr, Kazakova SV, Sambol SP, Johnson S, Gerding
DN. An epidemic, toxin gene-variant strain of Clostridium difficile. N
Engl J Med 2005; 353: 2433-41. https://doi.org/10.1056/NEJMoa051590
PMid:16322603

- Loo VG, Poirier L,
Miller MA, Oughton M, Libman MD, Michaud S, Bourgault AM, Nguyen T,
Frenette C, Kelly M, Vibien A, Brassard P, Fenn S, Dewar K, Hudson TJ,
Horn R, René P, Monczak Y, Dascal A. A predominantly clonal
multi-institutional outbreak of Clostridium difficile-associated
diarrhea with high morbidity and mortality. N Engl J Med 2005; 353:
2442-9. https://doi.org/10.1056/NEJMoa051639
PMid:16322602

- Teasley DG, Gerding
DN,
Olson MM, Peterson LR, Gebhard RL, Schwartz MJ, Lee JT Jr. Prospective
randomised trial of metronidazole versus vancomycin for Clostridium
difficile-associated diarrhoea and colitis. Lancet 1983; 2: 1043-6.
https://doi.org/10.1016/S0140-6736(83)91036-X

- Wenisch
C, Parschalk B, Hasenhündl M, Hirschl AM, Graninger W. Comparison of
vancomycin, teicoplanin, metronidazole, and fusidic acid for the
treatment of Clostridium difficile-associated diarrhea. Clin Infect Dis
1996; 22: 813-8. https://doi.org/10.1093/clinids/22.5.813
PMid:8722937

- Musher DM, Aslam S,
Logan N, Nallacheru S, Bhaila I, Borchert F, Hamill RJ. Relatively poor
outcome after treatment of Clostridium difficile colitis with
metronidazole. Clin Infect Dis 2005; 40: 1586-90.
https://doi.org/10.1086/430311
PMid:15889354

- Warny
M, Pepin J, Fang A, Killgore G, Thompson A, Brazier J, Frost E,
McDonald LC. Toxin production by an emerging strain of Clostridium
difficile associated with outbreaks of severe disease in North America
and Europe. Lancet 2005; 366: 1079-84.
https://doi.org/10.1016/S0140-6736(05)67420-X

- Al-Nassir
WN, Sethi AK, Nerandzic MM, Bobulsky GS, Jump RL, Donskey CJ.
Comparison of clinical and microbiological response to treatment of
Clostridium difficile-associated disease with metronidazole and
vancomycin. Clin Infect Dis 2008; 47: 56-62.
https://doi.org/10.1086/588293
PMid:18491964

- Zar
FA, Bakkanagari SR, Moorthi KM, Davis MB. A comparison of vancomycin
and metronidazole for the treatment of Clostridium difficile-associated
diarrhea, stratified by disease severity. Clin Infect Dis 2007; 45:
302-7. https://doi.org/10.1086/519265
PMid:17599306

- Schalk
E, Bohr UR, König B, Scheinpflug K, Mohren M. Clostridium
difficile-associated diarrhoea, a frequent complication in patients
with acute myeloid leukaemia. Ann Hematol 2010; 89: 9-14.
https://doi.org/10.1007/s00277-009-0772-0
PMid:19533126

- Gorschlüter M,
Glasmacher A, Hahn C, Schakowski F, Ziske C, Molitor E, Marklein G,
Sauerbruch T, Schmidt-Wolf IG. Clostridium difficile infection in
patients with neutropenia. Clin Infect Dis 2001; 33: 786-91.
https://doi.org/10.1086/322616
PMid:11512083

- Parmar
SR, Bhatt V, Yang J, Zhang Q, Schuster M. A retrospective review of
metronidazole and vancomycin in the management of Clostridium difficile
infection in patients with hematologic malignancies. J Oncol Pharm
Pract 2014; 20: 172-82. https://doi.org/10.1177/1078155213490004
PMid:23804627

- Cohen SH, Gerding DN,
Johnson S, Kelly CP, Loo VG, McDonald LC, Pepin J, Wilcox MH; Society
for Healthcare Epidemiology of America; Infectious Diseases Society of
America. Clinical practice guidelines for Clostridium difficile
infection in adults: 2010 update by the society for healthcare
epidemiology of America (SHEA) and the infectious diseases society of
America (IDSA). Infect Control Hosp Epidemiol 2010; 31: 431-55.
https://doi.org/10.1086/651706
PMid:20307191

- Clutter
DS, Dubrovskaya Y, Merl MY, Teperman L, Press R, Safdar A. Fidaxomicin
versus conventional antimicrobial therapy in 59 recipients of solid
organ and hematopoietic stem cell transplantation with Clostridium
difficile-associated diarrhea. Antimicrob Agents Chemother 2013; 57:
4501-5. https://doi.org/10.1128/AAC.01120-13
PMid:23836168
PMCid:PMC3754298

- Louie TJ, Miller MA,
Mullane KM, Weiss
K, Lentnek A, Golan Y, Gorbach S, Sears P, Shue YK; OPT-80-003 Clinical
Study Group. Fidaxomicin versus vancomycin for Clostridium difficile
infection. N Engl J Med 2011; 364: 422-31.
https://doi.org/10.1056/NEJMoa0910812
PMid:21288078

- Cornely
OA, Miller MA, Fantin B, Mullane K, Kean Y, Gorbach S. Resolution of
Clostridium difficile-associated diarrhea in patients with cancer
treated with fidaxomicin or vancomycin. J Clin Oncol 2013; 31: 2493-9.
https://doi.org/10.1200/JCO.2012.45.5899
PMid:23715579

- Cattaneo C, Casari S,
Bracchi F, Signorini L, Ravizzola G, Borlenghi E, Re A, Manca N, Carosi
G, Rossi G. Recent increase in enterococci, viridans streptococci,
Pseudomonas spp. and multiresistant strains among haematological
patients, with a negative impact on outcome. Results of a 3-year
surveillance study at a single institution. Scand J Infect Dis 2010;
42: 324-32. https://doi.org/10.3109/00365540903496569
PMid:20100118

- Haeusler GM, Mechinaud
F, Daley AJ, Starr M, Shann F, Connell TG, Bryant PA, Donath S, Curtis
N. Antibiotic-resistant gram-negative bacteremia in pediatric oncology
patients--risk factors and outcomes. Pediatr Infect Dis J 2013; 32:
723-6. https://doi.org/10.1097/INF.0b013e31828aebc8
PMid:23838774

- Ortega M, Marco F,
Soriano A,
Almela M, Martínez JA, Mu-oz A, Mensa J.Analysis of 4758 Escherichia
coli bacteraemia episodes: predictive factors for isolation of an
antibiotic-resistant strain and their impact on the outcome. J
Antimicrob Chemother 2009; 63: 568-74.
https://doi.org/10.1093/jac/dkn514
PMid:19126669

- Gyssens
IC, Kern W, Livermore DM. The Role of Antibiotic Stewardship in
Limiting Antibacterial Resistance for Haematology Patients. 4th
European Conference on Infections in Leukaemia. Meeting: September
8-10th, 2011. Final version: Feb 14th, 2012. Available at:
http://www.ebmt.org/Contents/Resources/Library/ECIL/Documents/Forms/AllItems.aspx
- Gudiol C, Carratalà J.
Antibiotic resistance in cancer
patients. Expert Rev Anti Infect Ther 2014; 12: 1003-16.
https://doi.org/10.1586/14787210.2014.920253
PMid:24834465

- Engelhard D, Akova M,
Boeckh MJ, et al. Bacterial infection prevention after hematopoietic
cell transplantation. Bone Marrow Transplant 2009; 44: 467-70.
https://doi.org/10.1038/bmt.2009.257
PMid:19861980

- Tomblyn
M, Chiller T, Einsele H, Gress R, Sepkowitz K, Storek J, et al.
Guidelines for preventing infectious complications among hematopoietic
cell transplantation recipients: A global perspective. Biol Blood
Marrow Transplant 2009; 15: 1143-1238.
https://doi.org/10.1016/j.bbmt.2009.06.019
PMid:19747629
PMCid:PMC3103296

- Boyce JM, Pittet D.
Guideline for hand
hygiene in health-care settings. Recommendations of the Healthcare
Infection Control Practices Advisory Committee and the
HICPAC/SHEA/APIC/IDSA Hand Hygiene Task Force. Society for Healthcare
Epidemiology of America/Association for Professionals in Infection
Control/Infectious Diseases Society of America. MMWR Recomm Rep 2002;
51(RR-16): 1-45, quiz CE1-4.

- Yokoe D, Casper C,
Dubberke
E, Lee G, Mu-oz P, Palmore T, Sepkowitz K, Young JA, Donnelly JP;
Center for International Blood and Marrow Transplant Research; National
Marrow Donor Program; European Blood and Marrow Transplant Group;
American Society of Blood and Marrow Transplantation; Canadian Blood
and Marrow Transplant Group; Infectious Disease Society of America;
Society for Healthcare Epidemiology of America; Association of Medical
Microbiology and Infectious Diseases Canada; Centers for Disease
Control and Prevention. Infection prevention and control in health-care
facilities in which hematopoietic cell transplant recipients are
treated. Bone Marrow Transplant 2009; 44: 495-507.
https://doi.org/10.1038/bmt.2009.261
PMid:19861984

- Sehulster
L, Chinn RY; CDC; HICPAC. Guidelines for environmental infection
control in health-care facilities. Recommendations of CDC and the
Healthcare Infection Control Practices Advisory Committee (HICPAC).
MMWR Recomm Rep 2003; 52(RR-10): 1-42. PMid:12836624

- Centers
for Disease Control and Prevention (CDC). Guidance for control of
infections with carbapenem-resistant or carbapenemase-producing
Enterobacteriaceae in acute care facilities. MMWR Morb Mortal Wkly Rep
2009; 58: 256-60. PMid:19300408

- Troché
G, Joly LM, Guibert M, Zazzo JF. Detection and treatment of
antibiotic-resistant bacterial carriage in a surgical intensive care
unit: a 6-year prospective survey. Infect Control Hosp Epidemiol 2005;
26: 161-5. https://doi.org/10.1086/502521
PMid:15756887

- Reddy P, Malczynski M,
Obias A, Reiner S, Jin N, Huang J, Noskin GA, Zembower T. Screening for
extended-spectrum beta-lactamase-producing Enterobacteriaceae among
high-risk patients and rates of subsequent bacteremia. Clin Infect Dis
2007; 45: 846-52. https://doi.org/10.1086/521260
PMid:17806048

- Gardam MA, Burrows LL,
Kus JV, Brunton J, Low DE, Conly JM, Humar A. Is surveillance for
multidrug-resistant enterobacteriaceae an effective infection control
strategy in the absence of an outbreak? J Infect Dis 2002; 186:
1754-60. https://doi.org/10.1086/345921
PMid:12447761

- Siegel JD, Rhinehart
E, Jackson M, Chiarello L;
Healthcare Infection Control Practices Advisory Committee. Management
of multidrug-resistant organisms in health care settings, 2006. Am J
Infect Control 2007; 35(10 Suppl 2): S165-93.
https://doi.org/10.1016/j.ajic.2007.10.006
PMid:18068814

- Yeh KM, Siu LK, Chang
JC, Chang FY. Vancomycin-resistant enterococcus (VRE) carriage and
infection in intensive care units. Microb Drug Resist 2004; 10: 177-83.
https://doi.org/10.1089/1076629041310091
PMid:15256034

- Muto CA, Giannetta ET,
Durbin LJ, Simonton BM, Farr BM. Cost-effectiveness of perirectal
surveillance cultures for controlling vancomycin-resistant
Enterococcus. Infect Control Hosp Epidemiol 2002; 23: 429-35.
https://doi.org/10.1086/502080
PMid:12186207

- Jernigan
JA, Titus MG, Gröschel DH, Getchell-White S, Farr BM. Effectiveness of
contact isolation during a hospital outbreak of methicillin-resistant
Staphylococcus aureus. Am J Epidemiol 1996; 143: 496-504.
https://doi.org/10.1093/oxfordjournals.aje.a008770
PMid:8610665

- Harbarth S, Fankhauser
C, Schrenzel J, Christenson J, Gervaz P, Bandiera-Clerc C, Renzi G,
Vernaz N, Sax H, Pittet D. Universal screening for
methicillin-resistant Staphylococcus aureus at hospital admission and
nosocomial infection in surgical patients. JAMA 2008; 299: 1149-57.
https://doi.org/10.1001/jama.299.10.1149
PMid:18334690

- Robicsek A, Beaumont
JL, Paule SM, Hacek DM, Thomson RB Jr, Kaul KL, King P, Peterson LR.
Universal surveillance for methicillin-resistant Staphylococcus aureus
in 3 affiliated hospitals. Ann Intern Med 2008; 148: 409-18.
https://doi.org/10.7326/0003-4819-148-6-200803180-00003
PMid:18347349

- Huang SS, Yokoe DS,
Hinrichsen VL, Spurchise LS, Datta R, Miroshnik I, Platt R.Impact of
routine intensive care unit surveillance cultures and resultant barrier
precautions on hospital-wide methicillin-resistant Staphylococcus
aureus bacteremia. Clin Infect Dis 2006; 43: 971-8.
https://doi.org/10.1086/507636
PMid:16983607

- Girmenia
C, Viscoli C, Piciocchi A, Cudillo L, Botti S, Errico A, Sarmati L,
Ciceri F, Locatelli F, Giannella M, Bassetti M, Tascini C, Lombardini
L, Majolino I, Farina C, Luzzaro F, Rossolini GM, Rambaldi A.
Management of carbapenem resistant Klebsiella pneumoniae infections in
stem cell transplant recipients: an Italian multidisciplinary consensus
statement. Haematologica. 2015; 100: e373-6.
https://doi.org/10.3324/haematol.2015.125484
PMid:25862702
PMCid:PMC4800687

- Metan G, Akova M.
Reducing the impact of
carbapenem-resistant Enterobacteriaceae on vulnerable patient groups:
what can be done? Curr Opin Infect Dis 2016; 29: 555-60.
https://doi.org/10.1097/QCO.0000000000000313
PMid:27584588

- Averbuch
D, Engelhard D. Gram-Negative Bacterial Infections After Hematopoietic
Stem Cell or Solid Organ Transplantation. In: Ljungman P, Snydman D,
Boeckh M (eds). Transplant Infections. Springer International
Publishing, Switzerland. 2016: 357-80.
https://doi.org/10.1007/978-3-319-28797-3_21

- Machuca I,
Gutiérrez-Gutiérrez B, Pérez Cortés S, Gracia-Ahufinger I, Serrano J,
Madrigal MD, Barcala J, Rodríguez-López F, Rodríguez-Ba-o J,
Torre-Cisneros J. Oral decontamination with aminoglycosides is
associated with lower risk of mortality and infections in high-risk
patients colonized with colistin-resistant, KPC-producing Klebsiella
pneumoniae. J Antimicrob Chemother 2016 Jul 26. pii: dkw272.
https://doi.org/10.1093/jac/dkw272

- Zuckerman T,
Benyamini N, Sprecher H, Fineman R, Finkelstein R, Rowe JM, Oren I. SCT
in patients with carbapenem resistant Klebsiella pneumoniae: a single
center experience with oral gentamicin for the eradication of carrier
state. Bone Marrow Transplant 2011; 46: 1226-30.
https://doi.org/10.1038/bmt.2010.279
PMid:21057549

- Saidel-Odes
L, Polachek H, Peled N, Riesenberg K, Schlaeffer F, Trabelsi Y, Eskira
S, Yousef B, Smolykov R, Codish S, Borer A. A randomized, double-blind,
placebo-controlled trial of selective digestive decontamination using
oral gentamicin and oral polymyxin E for eradication of
carbapenem-resistant Klebsiella pneumoniae carriage. Infect Control
Hosp Epidemiol 2012; 33: 14-9. https://doi.org/10.1086/663206
PMid:22173517

- Oren I, Sprecher H,
Finkelstein R, Hadad S, Neuberger A, Hussein K, Raz-Pasteur A, Lavi N,
Saad E, Henig I, Horowitz N, Avivi I, Benyamini N, Fineman R, Ofran Y,
Haddad N, Rowe JM, Zuckerman T. Eradication of carbapenem-resistant
Enterobacteriaceae gastrointestinal colonization with nonabsorbable
oral antibiotic treatment: A prospective controlled trial. Am J Infect
Control 2013; 41: 1167-72. https://doi.org/10.1016/j.ajic.2013.04.018
PMid:24274912

- Bar-Yoseph H, Hussein
K, Braun E, Paul M. Natural history and decolonization strategies for
ESBL/carbapenem-resistant Enterobacteriaceae carriage: systematic
review and meta-analysis. J Antimicrob Chemother 2016; 71: 2729-39.
https://doi.org/10.1093/jac/dkw221
PMid:27317444

- Lübbert
C, Faucheux S, Becker-Rux D, Laudi S, Dürrbeck A, Busch T, Gastmeier P,
Eckmanns T, Rodloff AC, Kaisers UX. Rapid emergence of secondary
resistance to gentamicin and colistin following selective digestive
decontamination in patients with KPC-2-producing Klebsiella pneumoniae:
a single-centre experience. Int J Antimicrob Agents 2013; 42: 565-70.
https://doi.org/10.1016/j.ijantimicag.2013.08.008
PMid:24100228

- Oostdijk
EA, Kesecioglu J, Schultz MJ, Visser CE, de Jonge E, van Essen EH,
Bernards AT, Purmer I, Brimicombe R, Bergmans D, van Tiel F, Bosch FH,
Mascini E, van Griethuysen A, Bindels A, Jansz A, van Steveninck FA,
van der Zwet WC, Fijen JW, Thijsen S, de Jong R, Oudbier J, Raben A,
van der Vorm E, Koeman M, Rothbarth P, Rijkeboer A, Gruteke P,
Hart-Sweet H, Peerbooms P, Winsser LJ, van Elsacker-Niele AM,
Demmendaal K, Brandenburg A, de Smet AM, Bonten MJ. Effects of
decontamination of the oropharynx and intestinal tract on antibiotic
resistance in ICUs: a randomized clinical trial. JAMA 2014; 312:
1429-37. https://doi.org/10.1001/jama.2014.7247
PMid:25271544

- Tascini C, Sbrana F,
Flammini S, Tagliaferri E, Arena F, Leonildi A, Ciullo I, Amadori F, Di
Paolo A, Ripoli A, Lewis R, Rossolini GM, Menichetti F; GENGUT Study
Group. Oral gentamicin gut decontamination for prevention of
KPC-producing Klebsiella pneumoniae infections: relevance of
concomitant systemic antibiotic therapy. Antimicrob Agents Chemother
2014; 58: 1972-6. https://doi.org/10.1128/AAC.02283-13
PMid:24419337
PMCid:PMC4023775

[TOP]




































































































































































