Reza Mirnejad1, Mohsen Heidary2*, Aghil Bahramian3, Mehdi Goudarzi3 and Abazar Pournajaf4.
1 Molecular
Biology Research Center, Systems Biology and Poisoning Institute,
Baqiyatallah University of Medical Sciences, Tehran, Iran.
2 Department of Microbiology, School of Medicine, Iran University of Medical Sciences, Tehran, Iran.
3 Department of Microbiology, School of Medicine, Shahid Beheshti University of Medical Sciences, Tehran, Iran.
4 Department of Microbiology, Faculty of Medicine, Babol University of medical sciences, Babol, Iran.
Corresponding
author: Mohsen Heidary, Department of Microbiology, School of Medicine,
Iran University of Medical Sciences, Tehran, Iran. E-mail:
mohsenheidary40@gmail.com
Published: July 1, 2018
Received: March 26, 2018
Accepted: May 15, 2018
Mediterr J Hematol Infect Dis 2018, 10(1): e2018044 DOI
10.4084/MJHID.2018.044
This article is available on PDF format at:

This is an Open Access article distributed
under the terms of the Creative Commons Attribution License
(https://creativecommons.org/licenses/by-nc/4.0),
which permits unrestricted use, distribution, and reproduction in any
medium, provided the original work is properly cited.
|
|
Abstract
Acinetobacter baumannii
is an important opportunistic pathogen, responsible for approximately
10% of all gram-negative nosocomial infection. The aim of this study
was to determine aminoglycoside and quinolone resistance genes and
their antimicrobial susceptibility profile in the clinically A. baumannii. In this cross-sectional study, a total of 100 nonduplicative A. baumannii
isolates were collected from different clinical samples. Antimicrobial
susceptibility test was performed by disk diffusion method. QnrA, anrB, qnrS, aac(3)-IIa, and aac(6′)-Ib
genes were identified using PCR method. The results of antibiotic
susceptibility test showed that polymyxin B was the most effective
antimicrobial against A. baumannii.
97%, 95% and 82% of isolates were resistant to cefepime, ceftriaxone,
and amikacin, respectively. The molecular distribution of aac(3)-IIa, aac(6′)-Ib, and qnrA genes were 45%, 50%, and 50% of isolates, respectively. However, qnrB and qnrS genes could not be detected in any strain. This study showed that polymyxin B was the best drug against A. baumannii clinical
isolates. This data is also valid for polymyxin E (colistin), which is
mostly used in clinics. There is a high level of resistance genes among
clinical A. baumannii
isolates. This high prevalence rate highlights the necessity for the
development of rapid diagnostic assays and continuous monitoring of
antibiotic resistance.
|
Introduction
Acinetobacter baumannii
is a lactose non-fermenting gram-negative bacillus (NF-GNB) that has
emerged as a highly troublesome pathogen particularly in critically ill
patients.[1] Clinical isolates of A. baumannii
are responsible for pulmonary, device-related, bloodstream, and urinary
tract infections and are frequently isolated from hospitalized ICU
patients.[2] These isolates were associated with multiple antibiotic resistance, and the spread of drug-resistant A. baumannii strains among hospitalized patients have become an increasing public health threat.[3,4]
Furthermore, due to the intrinsic resistance mechanisms in this
opportunistic nosocomial pathogen, it is quicker to become
multidrug-resistant (MDR).[5]
Polymyxin B and
polymyxin E (colistin), are an increasingly significant part of the
antimicrobial agents against MDR gram-negative bacteria. These two
drugs have the same spectrum and are appropriate for use in the
clinical settings.[6,7] At present in Europe in the patients with MDR A. baumannii the clinician utilize the colistin, and in the future, it is possible using the new derivatives.[8]
Aminoglycosides are used most commonly in the treatment of life-threatening infections caused by A. baumannii strains.[9]
The efflux pumps, decreased outer membrane permeability, amino acid
substitutions and enzymatic modification, are the main mechanisms of
aminoglycoside resistance in these bacteria.[10] Enzymatic modification is the most common type of aminoglycoside resistance in A. baumannii clinical isolates and usually results in high-level drug resistance.[11] Most enzyme-mediated resistance in A. baumannii
is due to the genes encoding for aminoglycoside-modifying enzymes
(AMEs) which found on plasmids and transposons. Three types of AMEs
include N-acetyltransferases (AAC), O-adenyltransferases (ANT), and
O-phosphotransferases (APH).[12,13]
The plasmid-mediated quinolone resistance (PMQR) genes, such as qnrA, qnrB, and qnrS, are responsible for quinolone resistance in A. baumannii isolates. PMQRs were first detected in the 1990s as a plasmid gene in Klebsiella pneumonia clinical isolates. Subsequent studies have shown that qnr
genes have a worldwide distribution in a range of Gram-negative
opportunist pathogens. Although the qnr expression mechanism which
confers clinical quinolone resistance is the least understood, the DNA
topoisomerase protection protein Qnr protects DNA from quinolone
binding and causes resistance to quinolones.[14-16] The prevalence of quinolone- and/or aminoglycoside-resistant A. baumannii
was increased during the past decade. The present study was carried out
to investigate antibiotic resistance pattern and resistance-related
genes such as qnrA, anrB, qnrS, aac(3)-IIa, and aac(6′)-Ib in A. baumannii clinical isolates by polymerase chain reaction (PCR) assay.
Materials and Methods
The
current study was a cross-sectional descriptive research which
conducted from February 2015 to April 2016, at two teaching hospitals
(Baqiyatallah and Moheb mehr hospitals) in Tehran, Iran. One hundred
non-repetitive strains of A. baumannii
were obtained from different clinical specimens, including tracheal
secretion, blood, wound, urine, and other samples. The isolates were
identified using well-recognized biochemical tests such as Gram
staining, oxidative/fermentative glucose test, catalase test, motility,
oxidase test, citrate utilization, and capability to grow at 42–44°C.[17] Species identification was confirmed by detection of blaOXA-51-like genes, as described previously.[18] All strains were preserved in Luria–Bertani broth (Merck Co., Germany) containing 20% glycerol (v/v) at –80°C for further use.
Antimicrobial
susceptibility was carried out on the Mueller-Hinton agar plates (Merck
Co., Germany) using the Kirby–Bauer (KB) method as suggested by the
Clinical and Laboratory Standards Institute guideline (CLSI document
M100-S14).
The antimicrobial agents were as follows: meropenem
(10 μg), gentamicin (10 μg), amikacin (30 μg), imipenem (10 μg),
tobramycin (10 μg), tetracycline (30 μg), piperacillin-tazobactam
(100-10 μg), cefepime (30 μg), ceftriaxone (30 μg),
ampicillin-sulbactam (10-10 μg), and polymyxin B (300 μg) (MAST
Diagnostics, Merseyside, UK). Multidrug-resistant (MDR), extensively
drug-resistant (XDR), and pan-drug-resistant (PDR) isolates were
detected according to the instruction suggested by the Centers for
Disease Control and Prevention (CDC). Escherichia coli ATCC 25922 and Acinetobacter baumannii ATCC 19606 were used as negative and positive controls, respectively.
Genomic DNA was extracted from A. baumannii colonies
grown overnight on blood agar by Bioneer Co., Korea Kit and used as a
template for PCR assay. PCR amplification was done to detect
aminoglycoside-(aac(3)-IIa and aac(6′)-Ib) and quinolone-(qnrA, qnrB, and qnrS)
related resistance genes. Amplification of AME and PMQR genes was
carried out using a thermal gradient cycler (Eppendorf Co., Germany)
with the following protocol: 5 minutes at 94°C for the initial
denaturation and 36 cycles of amplification consisting of 45 seconds at
94°C, 45 seconds at 52-58°C, and 45 seconds at 72°C, with 5 minutes at
72°C for the final extension. The specific primers, temperatures of
annealing, and amplicons size used for PCR are detailed in Table 1.
The
current survey was a descriptive research. The MINITAB16 software was
used for statistical analyses. The P value and confidence intervals were
≤0.05 and 95%, respectively.
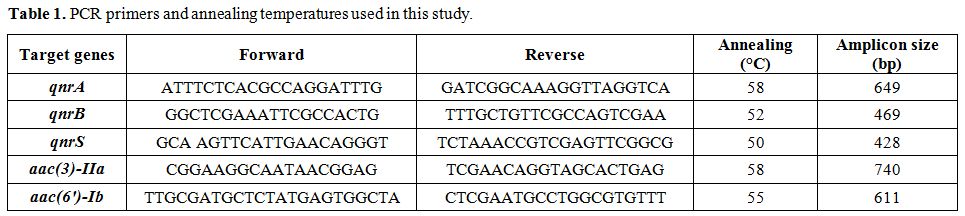 |
Table 1. PCR primers and annealing temperatures used in this study. |
Results
One hundred isolates of A. baumannii
were obtained from different clinical specimens. The samples included
blood (n=40, 40%), tracheal secretion (n=27, 27%), wound (n=12, 12%),
urine (n=8, 8%), and unknown (n=13, 13%) specimens isolated from
hospitalized patients in ICU (n=40, 40%), emergency department (n=20,
20%), and infectious disease department (30, 30%), and other
departments (n=10, 10%).
The resistance percentage of meropenem,
gentamicin, amikacin, imipenem, tobramycin, tetracycline,
piperacillin-tazobactam, cefepime, ceftriaxone, ampicillin-sulbactam
and polymyxin B were 69%, 82%, 63%, 74%, 56%, 51%, 70%, 97%, 95%, 49%,
3%, respectively (Table 2).
Antibiotic
susceptibility tests using the Kirby-Bauer method showed that the level
of resistance to meropenem, gentamicin, amikacin, imipenem, tobramycin,
tetracycline, piperacillin-tazobactam, cefepime, ceftriaxone,
ampicillin-sulbactam and polymyxin B was 69%, 82%, 63%, 74%, 56%, 51%,
70%, 97%, 95%, 49%, 3% (Table 2).
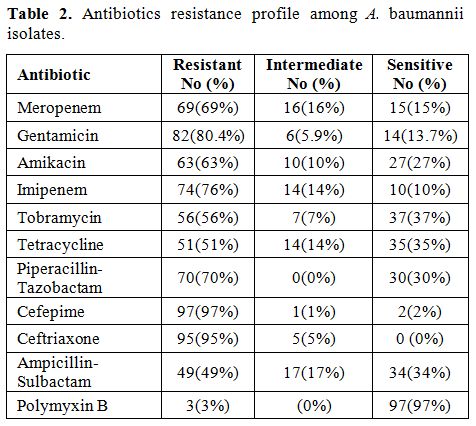 |
Table
2. Antibiotics resistance profile among A. baumannii isolates. |
All 100 isolates of the main outbreak strains of A. baumannii were PCR positive for blaOXA-51-like genes.
Molecular distribution of aminoglycoside resistance genes including aac(3)-IIa and aac(6′)-Ib were 45% and 50%, respectively is shown in the figure 1 A and B. Half of the isolates (50%) contained the qnrA (Figure 1C). QnrB and qnrS were not found in any strains. Sequencing of PCR products for AME and PMQR genes were confirmed by BLAST at NCBI.
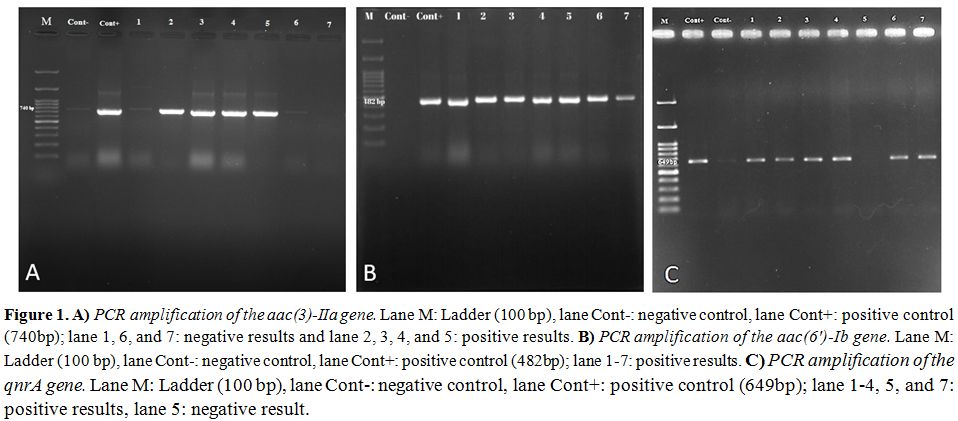 |
Figure 1. A) PCR amplification of the
aac(3)-IIa gene. Lane M: Ladder (100 bp), lane Cont-: negative control,
lane Cont+: positive control (740bp); lane 1, 6, and 7: negative
results and lane 2, 3, 4, and 5: positive results. B) PCR amplification
of the aac(6′)-Ib gene. Lane M: Ladder (100 bp), lane Cont-: negative
control, lane Cont+: positive control (482bp); lane 1-7: positive
results. C) PCR amplification of the qnrA gene. Lane M: Ladder (100
bp), lane Cont-: negative control, lane Cont+: positive control
(649bp); lane 1-4, 5, and 7: positive results, lane 5: negative result. |
Discussion
Drug resistance in A. baumannii
has become a global problem for the severely infected patients who
critically rely on Antimicrobial therapy. The emergence of clinical A. baumannii strains with different antibiotic resistance phenotypes causes difficulties in treating infections caused by this organism.[19,20]
Multidrug-resistant A. baumannii
(MDR-Ab) is a subject of profound anxiety as it not only causes severe
and fatal infections but also increases the length of hospital stay,
resulting in augmented treatment charges.[21]
In this study, the most antibiotic resistance in A. baumannii
isolates were related to cefepime (97%), ceftriaxone (95%), and
amikacin (82%), and the most effective drug against these isolates was
polymyxin B. This data is also valid for colistin, which is mostly used
in clinics worldwide.
Henwood et al.,[22] showed that more than 75% of A. baumannii strains were resistant to cefotaxime and ceftazidime. In another study, Karlowsky et al.,[23] showed that >90% of A. baumannii
isolates were susceptible to imipenem and meropenem; fewer strains were
susceptible to amikacin, and <60% were susceptible to ceftazidime
and gentamicin.
In agreement with the current study, polymyxins, are active agents against the overwhelming majority of A. baumannii throughout the world. In a systematic review study directed by Razavi Nikoo et al.,[24] polymyxins presented adequate activity against A. baumannii
collected. The frequencies of MDR and XDR isolates were 70% and 19%
respectively. No PDR isolates were identified in this study.
Hujer et al.[25] in their study reported that 89% of A. baumannii were resistant to at least three different classes of antibiotics, and 15% were resistant to all antibiotics tested.
Aminoglycosides are used most commonly in the treatment of A. baumannii infections. Most enzyme-mediated resistance in A. baumannii is due to AMEs encoded genes which found on the mobile genetic elements.
PMQR genes including qnrA, qnrB, and qnrS are responsible for quinolone resistance in A. baumannii which the prevalence of quinolone-resistant A. baumannii was increased in recent years. In our study, the prevalence rate of PMQR genes including qnrA, qnrB, and qnrS was 50%, 0%, and 0%, respectively. In contrast with our data, Chagas et al.,[26] showed that the prevalence of qnrA
gene was 37.5% (n=15). The differences mentioned above can result from
the geographical distance, surveillance strategies, and restraint in
antibiotic prescriptions in other regions.
Conclusions
This study showed that the most effective antibiotic against clinical strains of A. baumannii was polymyxin B and we recommend clinicians to use polymyxins (B or E) in patients infected with MDR A. baumannii.
However, overusing can lead to polymyxin resistance, and the drug's
toxicity problems should be considered. There is a high level of
aminoglycoside resistance genes among A. baumannii isolates circulating in hospitals in Iran. This trend of MDR profiles associated with the presence of aac(6′)-Ib and aac(3)-IIa genes
are worrying. The high prevalence rate of these resistance genes
highlights the necessity for establishing more rapid diagnostic assays,
more antimicrobial susceptibility tests, more clinician-laboratory
correlation, and continuous monitoring of antibiotic resistance due to A. baumannii.
References
- Juyal D, Prakash R, Shanakarnarayan SA, Sharma M,
Negi V, Sharma N. Prevalence of non-fermenting gram negative bacilli
and their in vitro susceptibility pattern in a tertiary care hospital
of Uttarakhand: A study from foothills of Himalayas. Prevalence.
2013;2(2):108-12. https://doi.org/10.4103/2278-0521.117915

- Almasaudi
SB. Acinetobacter spp. as nosocomial pathogens: Epidemiology and
resistance features. Saudi Journal of Biological Sciences. 2016.

- Dijkshoorn
L, Nemec A, Seifert H. An increasing threat in hospitals:
multidrug-resistant Acinetobacter baumannii. Nature Reviews
Microbiology. 2007;5(12):939. https://doi.org/10.1038/nrmicro1789 PMid:18007677

- Turton
JF, Woodford N, Glover J, Yarde S, Kaufmann ME, Pitt TL. Identification
of Acinetobacter baumannii by detection of the blaOXA-51-like
carbapenemase gene intrinsic to this species. Journal of Clinical
Microbiology. 2006;44(8):2974-6. https://doi.org/10.1128/JCM.01021-06 PMid:16891520 PMCid:PMC1594603

- Gholami
M, Haghshenas M, Moshiri M, Razavi S, Pournajaf A, Irajian G, et al.
Frequency of 16S rRNA Methylase and Aminoglycoside-Modifying Enzyme
Genes among Clinical Isolates of Acinetobacter baumannii in Iran.
Iranian Journal of Pathology. 2017;12(4):329.

- Cai Y, Lee W, Kwa AL. Polymyxin B versus colistin: an update. Expert review of anti-infective therapy. 2015;13(12):1481-97. https://doi.org/10.1586/14787210.2015.1093933 PMid:26488563

- Kassamali
Z, Danziger L. To B or not to B, that is the question: is it time to
replace colistin with polymyxin B? Pharmacotherapy: The Journal of
Human Pharmacology and Drug Therapy. 2015;35(1):17-21. https://doi.org/10.1002/phar.1510 PMid:25346395

- Vaara
M. New polymyxin derivatives that display improved efficacy in animal
infection models as compared to polymyxin B and colistin. Medicinal
Research Reviews. 2018. https://doi.org/10.1002/med.21494 PMid:29485690

- Viehman
JA, Nguyen MH, Doi Y. Treatment options for carbapenem-resistant and
extensively drug-resistant Acinetobacter baumannii infections. Drugs.
2014;74(12):1315-33. https://doi.org/10.1007/s40265-014-0267-8 PMid:25091170 PMCid:PMC4258832

- Potron
A, Poirel L, Nordmann P. Emerging broad-spectrum resistance in
Pseudomonas aeruginosa and Acinetobacter baumannii: mechanisms and
epidemiology. International journal of antimicrobial agents.
2015;45(6):568-85. https://doi.org/10.1016/j.ijantimicag.2015.03.001 PMid:25857949

- Kashfi
M, Hashemi A, Eslami G, Amin MS, Tarashi S, Taki E. The Prevalence of
Aminoglycoside-Modifying Enzyme Genes Among Pseudomonas aeruginosa
Strains Isolated From Burn Patients. Archives of Clinical Infectious
Diseases. 2017;12(1).

- Huang
L, Sun L, Yan Y. Time to positivity of blood culture is predictive for
nosocomial infection and infectious endocarditis instead of other
clinical characteristics and prognosis in Acintobacter baumannii
bloodstream infection. Journal of Infection. 2014;68(2):198-200. https://doi.org/10.1016/j.jinf.2013.10.004 PMid:24140064

- Khoshnood
S, Eslami G, Hashemi A, Bahramian A, Heidary M, Yousefi N, et al.
Distribution of Aminoglycoside Resistance Genes Among Acinetobacter
baumannii Strains Isolated From Burn Patients in Tehran, Iran. Archives
of Pediatric Infectious Diseases. 2017;5(3). https://doi.org/10.5812/pedinfect.57263

- Xiong
X, Bromley EH, Oelschlaeger P, Woolfson DN, Spencer J. Structural
insights into quinolone antibiotic resistance mediated by pentapeptide
repeat proteins: conserved surface loops direct the activity of a Qnr
protein from a Gram-negative bacterium. Nucleic Acids Research.
2011;39(9):3917-27. https://doi.org/10.1093/nar/gkq1296 PMid:21227918 PMCid:PMC3089455

- Robicsek
A, Jacoby GA, Hooper DC. The worldwide emergence of plasmid-mediated
quinolone resistance. The Lancet Infectious Diseases.
2006;6(10):629-40. https://doi.org/10.1016/S1473-3099(06)70599-0

- Yang
HY, Nam YS, Lee HJ. Prevalence of plasmid-mediated quinolone resistance
genes among ciprofloxacin-nonsusceptible Escherichia coli and
Klebsiella pneumoniae isolated from blood cultures in Korea. Canadian
Journal of Infectious Diseases and Medical Microbiology.
2014;25(3):163-9. https://doi.org/10.1155/2014/329541 PMid:25285114 PMCid:PMC4173980

- Camp
C, Tatum OL. A review of Acinetobacter baumannii as a highly successful
pathogen in times of war. Laboratory Medicine. 2015;41(11):649-57. https://doi.org/10.1309/LM90IJNDDDWRI3RE

- Leski
TA, Bangura U, Jimmy DH, Ansumana R, Lizewski SE, Li RW, et al.
Identification of blaOXA-51-like, blaOXA-58, blaDIM-1, and blaVIM
carbapenemase genes in hospital Enterobacteriaceae isolates from
Sierra Leone. Journal of Clinical Microbiology.
2013;51(7):2435-8. https://doi.org/10.1128/JCM.00832-13 PMid:23658259 PMCid:PMC3697688

- Antunes
L, Visca P, Towner KJ. Acinetobacter baumannii: evolution of a global
pathogen. Pathogens and Disease. 2014;71(3):292-301. https://doi.org/10.1111/2049-632X.12125 PMid:24376225

- Babapour
E, Haddadi A, Mirnejad R, Angaji S-A, Amirmozafari N. Study of drug
resistance and ompA gene existence in clinical Acinetobacter baumannii
isolates. Iran J Med Microbiol 2017, 11(1): 30-38

- Zarrilli
R, Pournaras S, Giannouli M, Tsakris A. Global evolution of
multidrug-resistant Acinetobacter baumannii clonal lineages.
International Journal of Antimicrobial Agents. 2013;41(1):11-9. https://doi.org/10.1016/j.ijantimicag.2012.09.008 PMid:23127486

- Henwood
CJ, Gatward T, Warner M, James D, Stockdale MW, Spence RP, et al.
Antibiotic resistance among clinical isolates of Acinetobacter in the
UK, and in vitro evaluation of tigecycline (GAR-936). Journal of
Antimicrobial Chemotherapy. 2002;49(3):479-87. https://doi.org/10.1093/jac/49.3.479 PMid:11864948

- Karlowsky
JA, Draghi DC, Jones ME, Thornsberry C, Friedland IR, Sahm DF.
Surveillance for antimicrobial susceptibility among clinical isolates
of Pseudomonas aeruginosa and Acinetobacter baumannii from hospitalized
patients in the United States, 1998 to 2001. Antimicrobial Agents and
Chemotherapy. 2003;47(5):1681-8. https://doi.org/10.1128/AAC.47.5.1681-1688.2003 PMid:12709340 PMCid:PMC153334

- Razavi
Nikoo H, Ardebili A, Mardaneh J. Systematic review of antimicrobial
resistance of clinical Acinetobacter baumannii isolates in Iran: an
update. Microbial Drug Resistance. 2017;23(6):744-56. https://doi.org/10.1089/mdr.2016.0118

- Hujer
KM, Hujer AM, Hulten EA, Bajaksouzian S, Adams JM, Donskey CJ, et al.
Analysis of antibiotic resistance genes in multidrug-resistant
Acinetobacter sp. isolates from military and civilian patients treated
at the Walter Reed Army Medical Center. Antimicrobial Agents and
Chemotherapy. 2006;50(12):4114-23. https://doi.org/10.1128/AAC.00778-06 PMid:17000742 PMCid:PMC1694013

- Chagas
TPG, Oliveira TRT, Ribeiro SS, Aires CAM, Carvalho-Assef, APD’A, Asensi
MD. Detection of plasmid-mediated Quinolone resistance genes (qnr)
among Acinetobacter baumannii isolated from BRAZIL. XXVIII Congresso
Brasileiro Microbiologia 2016. https://www.researchgate.net/publication/318223832https://www.researchgate.net/publication/318223832
[TOP]




























