Andreas Giannopoulos1, Niki Rougkala1,2, Theodoros Loupis1, Marina Mantzourani2, Nora-Athina Viniou2, Eleni Variami2, Theodoros P. Vassilakopoulos3, George Dryllis1,3, Ioannis Kotsianidis4, Theodora Gougopoulou5, Marianna Politou6, Kostas Konstantopoulos3 and George Vassilopoulos1,7.
1 Haematology Research Laboratory, Biomedical Research Foundation, Academy of Athens, Athens, Greece.
2 1st Department of Internal Medicine, Laikon Hospital, School of Medicine, University of Athens, Athens, Greece.
3
Department of Haematology and Bone Marrow Transplantation, Laikon
Hospital, School of Medicine, University of Athens, Athens, Greece.
4 Department of Haematology, Democritus University of Thrace, Medical School, Alexandroupolis, Greece.
5 Haematology
Clinic, Department of Internal Medicine, Faculty of Medicine, School of
Health Sciences, University of Ioannina, Ioannina, Greece.
6
Laboratory of Haematology and Blood Transfusion Unit, Aretaieion
Hospital, School of Medicine, University of Athens, Athens, Greece.
7 Department of Haematology, University of Thessaly Medical School, Larissa, Greece.
Correspondence to: George Vassilopoulos. MD PhD. Professor and Head,
Department of Hematology, Larisa University Hospital, Biopolis 1,
Larisa, GR41110. FAX +30 241350 1625. E-mail:
gvasilop@med.uth.gr
Published: January 1, 2019
Received: August 30, 2018
Accepted: December 14, 2018
Mediterr J Hematol Infect Dis 2019, 11(1): e2019009 DOI
10.4084/MJHID.2019.009
This is an Open Access article distributed
under the terms of the Creative Commons Attribution License
(https://creativecommons.org/licenses/by-nc/4.0),
which permits unrestricted use, distribution, and reproduction in any
medium, provided the original work is properly cited.
|
|
Abstract
Background and Objectives: Somatic mutations in the calreticulin gene (CALR)
are detected in approximately 70% of patients with essential
thrombocythemia (ET) and primary or secondary myelofibrosis (MF),
lacking the JAK2 and MPL mutations. To determine the prevalence of CALR
frameshift mutations in a population of MPN patients of Greek origin,
we developed a rapid low-budget PCR-based assay and screened samples
from 5 tertiary Haematology units. This is a first of its kind report
of the Greek patient population that also disclosed novel CALR mutants.
Methods: MPN patient samples were collected from different clinical units and screened for JAK2 and MPL mutations after informed consent was obtained. Negative samples were analyzed for the presence of CALR
mutations. To this end, we developed a modified post Real Time PCR
High-Resolution Melting Curve analysis (HRM-A) protocol. Samples were
subsequently confirmed by Sanger sequencing.
Results: Using this protocol we screened 173 MPN, JAK2 and MPL mutation negative, patients of Greek origin, of whom 117 (67.63%) displayed a CALR exon
nine mutation. More specifically, mutations were detected in 90 out of
130 (69.23%) essential thrombocythaemia cases (ET), in 18 out of 33
(54.55%) primary myelofibrosis patients (pMF) and in 9 out of 10 (90%)
cases of myelofibrosis secondary to ET (post-ET sMF). False positive
results were not detected. The limit of detection (LoD) of our protocol
was 2%. Furthermore, our study revealed six rare novel mutations which
are to be added in the COSMIC database.
Conclusions:
Overall, our method could rapidly and cost-effectively detect the
mutation status in a representative cohort of Greek patients; the
mutation make-up in our group was not different from what has been
published for other national groups.
|
Introduction
Philadelphia
chromosome-negative myeloproliferative neoplasms (MPNs) include
polycythaemia vera (PV), essential thrombocythaemia (ET) and primary
myelofibrosis (pMF).[1] The molecular basis of these disorders was partly elucidated in 2005 with the identification of the JAK2V617F mutation in the majority of PV patients and in about 60% of those with ET and MF.[2-6] It was later reported that somatic mutations of JAK2 exon 12 are present in the remaining PV patients,[7] while mutations of MPL exon 10 were present in about 5% of the ET and MF cases.[8] 2008 WHO definition of MPNs includes JAK2V617F or MPL exon ten mutations as a major criterion for disease diagnosis.[9] Beyond these two culprits, somatic mutations in the CALR gene that encodes for calreticulin were identified in 2013 in about 20-25% of patients with ET and MF[10,11] and were subsequently incorporated into the 2016 WHO diagnostic criteria.[12] Determination of CALR mutations is relevant not only for their diagnostic contribution[12] but also for their prognostic significance as well.[13-16]
CALR mutations
are deletion or insertion events or a combination of both within the
DNA sequence of the last exon (exon 9) of the gene. The two most common
mutations are either a 52-bp deletion (Type-1;
c.1099_1150del;L367fs*46; 45-55% of the cases) or a 5-bp insertion
(Type-2; c.1154_1155insTTGTC; K385fs*47; 32-42% of the cases). The
remaining 15% of the cases comprise other deletions or insertions or a
combination of both that are either unique or found in a small number
of patients.[10,11,17] All CALR
mutations result in a +1bp frameshift, leading to the coding of a novel
amino acid peptide sequence distal to the site of the mutation that
consequently generates a novel C- terminus at the protein level.[10,11]
The high incidence and specificity of CALR
mutations in ET and MF make inevitable the need of incorporating rapid
and sensitive methodologies in the diagnostic work-up of MPNs. In the
relatively similar case of JAK2, improved PCR methods, have pushed the JAK2V617F
mutation limit of detection (LoD) down to a burden of only 1-3%, which
has been considered sufficient for the clinical correlation to PV. Such
LoD levels should also be attained with the available technologies in
the case of CALR.[18]
Detection of CALR
exon nine mutations can be achieved by several methods such as Sanger
sequencing, fragment analysis, high resolution melting curve analysis
(HRM-A), TaqMan-based Real-Time PCR and targeted next-generation
sequencing (NGS).[10,11,19-22] Targeted NGS provides the best LoD and is the most robust technique;[21]
however, it is still a costly approach with a long turn-around time,
which makes it impractical for routine diagnostic services. HRM-A, is a
well-established method for the detection of gene polymorphisms and
mutations, by measuring changes in the melting point of DNA duplex.[23]
In this report, we describe a rapid and sensitive HRM-A protocol, which we developed for the detection of CALR
exon nine mutations using the LightCycler cobas 4800 platforms (ROCHE
Diagnostics, Indianapolis, IN, US). Our assay was first applied in a
small cohort of MF patients; subsequently, when thoroughly validated,
it was used for the study of ET and MF samples from several tertiary
health care centres in Greece, thus providing reliable data concerning
the frequency of CALR mutations within the Greek population and even disclosing a few novel mutations.
Material and Methods
Patient diagnosis was based on the WHO 2008 criteria.[1,9] CALR mutational analysis was performed in 130 patients with ET and 43 patients with MF who were negative for the JAK2V617F and MPLW515L/K mutations, as well as in 19 patients with secondary thrombocytosis.
The
study protocol was approved by the Internal Review Boards of all
participating Institutes; written informed consent was obtained from
all patients, and the study was conducted in accordance with the
current version of the Helsinki Declaration.
DNA was extracted by
standard procedures after isolation of total leukocytes from peripheral
blood or bone marrow following red cell lysis. All samples were
investigated for the presence of the JAK2V617F and the MPLW515L/K mutations. The JAK2V617F
mutation was detected using a tetra-primer amplification refractory
mutation system (ARMS) polymerase chain reaction (PCR) assay with a
sensitivity of 1% as previously described.[4] The MPLW515L/K mutations were detected using an allele-specific PCR (AS-PCR) assay with a sensitivity of 1% as previously described.[24]
HRM analysis assay design.
The HRM analysis assay was performed using the LightCycler cobas 4800
platform (ROCHE Diagnostics, Indianapolis, IN, US). Oligonucleotide
primers were designed using the Oligo7 Primer Analysis Software
v.7.0.5.7 (Molecular Biology Insights Inc, Colorado Springs, CO, US) to
flank all CALR exon 9 variants reported in MPNs. Primer sequences were
CALRe9HF: 5’- AGGCAGCAGAGAAACAAATGAA-3’ and CALRe9HR:
5’-TCTACAGCTCGTCCTTGGC-3’, and the amplicon size was 204 bp (GenBank:
NG_029662.1). Ten nanograms of DNA was amplified in a final volume of
10μL containing 1x LightCycler 480 High-Resolution Melting Master Mix
(ROCHE Diagnostics, Indianapolis, IN, US), 1.0μΜ of the CALRe9HF, 0.2μΜ
of the CALRe9HR and 2.5 mM MgCl2.
The
cycling conditions were: initial denaturation at 95°C for 10 min,
followed by 45 cycles of 95°C for 15s, 67°C for 30s and 72°C for 15s.
The high resolution melting program included denaturation at 95°C for 1
min, re-naturation at 40°C for 1 min and melting from 65oC to 95°C,
with a ramp of 0.03°C per second and 17 fluorescent acquisitions per
degree centigrade.
All samples were analysed in duplicate. Two samples of normal individuals (CALR wild-type), one positive control for CALR Type-1 and one positive control for CALR
Type-2 were included in each experiment. The analysis was done with the
Light Cycler 480 Gene Scanning Software v.1.5.1.74 (ROCHE Diagnostics,
Indianapolis, IN, US). Melting profiles were normalized, grouped and
displayed as fluorescence versus temperature plots. Normalization bars
were set between 81.5 and 82.0°C for the leading range and 88.0-88.5°C
for the tailing range. The threshold for the melting temperature (Tm)
shift was set at 10. The settings were optimized to a sensitivity value
of 0.4 on the analysis software.
Wild-type and mutated samples
were defined as negative and positive controls respectively in the
analysis. Melting curve analysis is based on the differences in melting
curve shape of each sample, hence clustering the samples into groups
based on the internal software calculation.
Sanger Sequencing. All patients were screened for CALR
exon 9 mutations spanning codons 1054-1254 by Sanger sequencing at
an ABI 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA, US).
Sanger
sequencing was performed on a broader genomic area than the one used
for the HRM-A. The PCR primer sequences were CALRe9SF: 5’-
CCAACGATGAGGCATACGCT-3’ and CALRe9SR: 5’-ATCCACCCCAAATCCGAACC-3’ and
the amplicon size was 469 bp (GenBank: NG_029662.1). PCR products were
cleaned using the QiaQuick Gel Extraction kit (Qiagen) according to the
manufacturer’s instructions. Bidirectional sequencing was performed
using the BigDye Terminator, v.3.1 Cycle Sequencing kit (Applied
Biosystems, Foster City, CA). The analysis was performed using the
Chromas v.2.4.3 (Technelysium Pty Ltd, South Brisbane, QLD, AU)
software. Results
HRM analysis for CALR exon 9 mutations. Table 1 summarizes our results. Of the 173 ET and MF patients analysed using our HRM-A protocol, 117 (67.63%) displayed a CALR
exon 9 mutation; the incidence of this finding in the ET and MF
cohorts was quite similar, comprising of 69.23% in the ET (90 out of
130) and 62.79% in the MF (27 out of 43) group of patients. More
specifically, the latter group includes 33 pMF and 10 secondary
post-ET MF, of which 54.55% (18 out of 33) and 90% (9 out of 10)
carried CALR exon 9 mutations, respectively. In contrast, all 19 patients with secondary thrombocytosis were CALR negative.
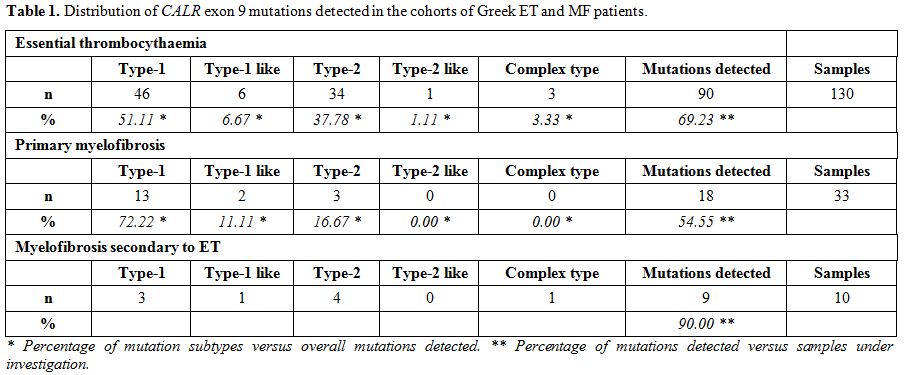 |
Table 1. Distribution of CALR exon 9 mutations detected in the cohorts of Greek ET and MF patients. |
Of the 90 ET
patients with HRM positive curves, 46 were Type-1 (L367fs*46), 6 were
Type-1-like (E364fs*55, L367fs*50, L367fs*52, D373fs*47, K375fs*49 and
K377fs*47), 34 were Type-2 (K385fs*47), 1 was Type-2-like (E386fs*46)
and 3 showed complex mutations consisting of D373fs*56, D373fs*51 and
K377fs*55. Of the 27 MF patients with HRM positive curves, 16 were
Type-1 (L367fs*46), 3 were Type-1-like (L367fs*52 and K377fs*47), 7
were Type-2 (K385fs*47) and 1 showed a complex mutation (E379fs*47).
Table 2 shows the coding sequence alterations (at the cDNA level) of the detected CALR
exon 9 mutations. Most of them are already known and included in the
most recent database of the Catalogue of Somatic Mutations in Cancer
(COSMIC v85, as of May 8th, 2017),[25] while six are reported for the first time. As expected, most commonly found mutations were those of Type-1 and Type-2 (Figure 1A).
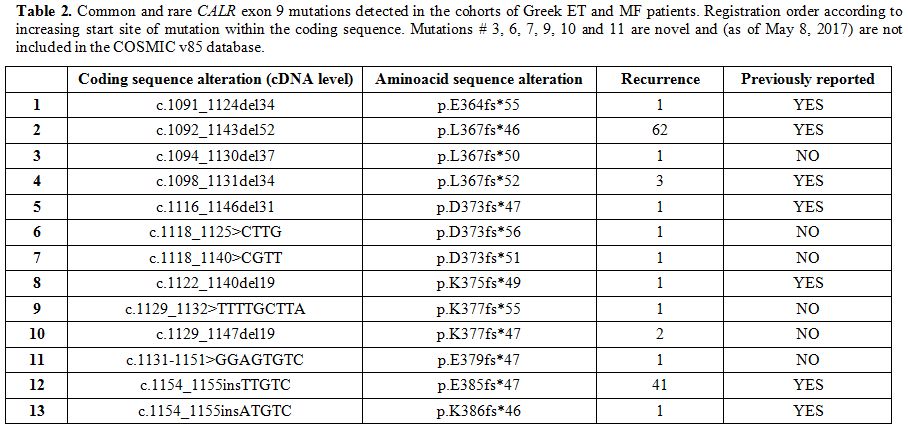 |
Table 2. Common and rare CALR exon 9
mutations detected in the cohorts of Greek ET and MF patients.
Registration order according to increasing start site of mutation
within the coding sequence. Mutations # 3, 6, 7, 9, 10 and 11 are novel
and (as of May 8, 2017) are not included in the COSMIC v85 database. |
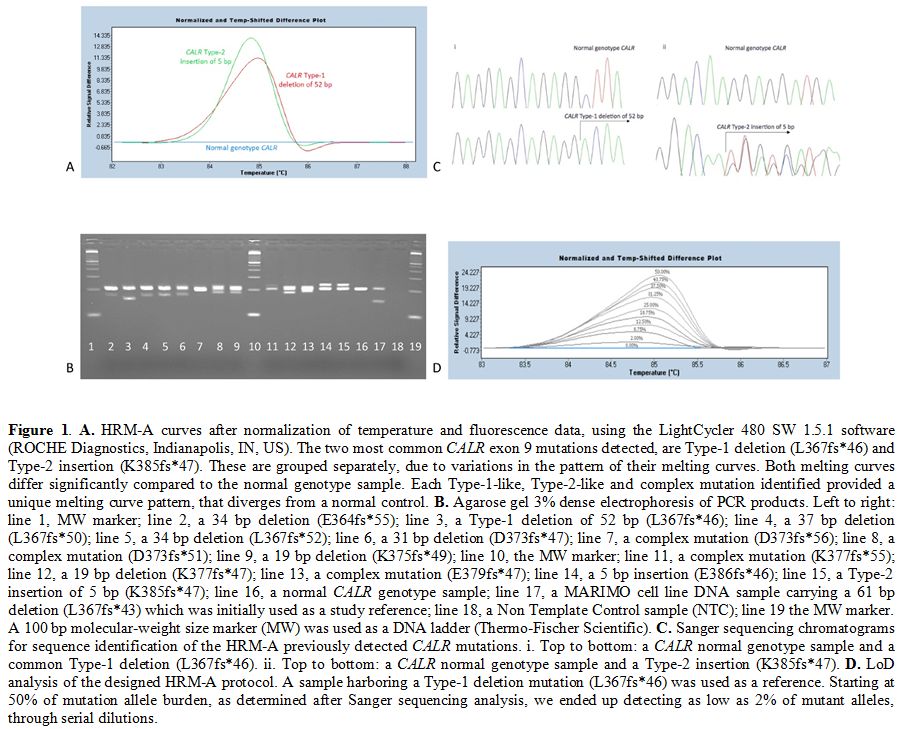 |
Figure 1. A.
HRM-A curves after normalization of temperature and fluorescence data,
using the LightCycler 480 SW 1.5.1 software (ROCHE Diagnostics,
Indianapolis, IN, US). The two most common CALR exon 9 mutations
detected, are Type-1 deletion (L367fs*46) and Type-2 insertion
(K385fs*47). These are grouped separately, due to variations in the
pattern of their melting curves. Both melting curves differ
significantly compared to the normal genotype sample. Each Type-1-like,
Type-2-like and complex mutation identified provided a unique melting
curve pattern, that diverges from a normal control. B.
Agarose gel 3% dense electrophoresis of PCR products. Left to right:
line 1, MW marker; line 2, a 34 bp deletion (E364fs*55); line 3, a
Type-1 deletion of 52 bp (L367fs*46); line 4, a 37 bp deletion
(L367fs*50); line 5, a 34 bp deletion (L367fs*52); line 6, a 31 bp
deletion (D373fs*47); line 7, a complex mutation (D373fs*56); line 8, a
complex mutation (D373fs*51); line 9, a 19 bp deletion (K375fs*49);
line 10, the MW marker; line 11, a complex mutation (K377fs*55); line
12, a 19 bp deletion (K377fs*47); line 13, a complex mutation
(E379fs*47); line 14, a 5 bp insertion (E386fs*46); line 15, a Type-2
insertion of 5 bp (K385fs*47); line 16, a normal CALR genotype sample;
line 17, a MARIMO cell line DNA sample carrying a 61 bp deletion
(L367fs*43) which was initially used as a study reference; line 18, a
Non Template Control sample (NTC); line 19 the MW marker. A 100 bp
molecular-weight size marker (MW) was used as a DNA ladder
(Thermo-Fischer Scientific). C.
Sanger sequencing chromatograms for sequence identification of the
HRM-A previously detected CALR mutations. i. Top to bottom: a CALR
normal genotype sample and a common Type-1 deletion (L367fs*46). ii.
Top to bottom: a CALR normal genotype sample and a Type-2 insertion
(K385fs*47). D. LoD analysis
of the designed HRM-A protocol. A sample harboring a Type-1 deletion
mutation (L367fs*46) was used as a reference. Starting at 50% of
mutation allele burden, as determined after Sanger sequencing analysis,
we ended up detecting as low as 2% of mutant alleles, through serial
dilutions. |
Following
HRM-A, PCR products were analysed in 3% agarose gel to confirm that the
detected melting curve pattern, diverging from normal genotype control
samples, actually belongs to a CALR exon 9 mutation (Figure 1B). The 204 bp band corresponds to the wild- type CALR gene, while additional bands are evidence of CALR mutations.
Sanger sequencing analysis of CALR
exon 9 mutations. Sanger sequencing analysis was performed in all
cases; results were fully concordant with those obtained by the HRM-A
technique (Figure 1C).
Limit of Detection of HRM analysis for CALR exon 9 mutations. In order to assess the LoD of the assay, we performed serial dilutions of a patient’s sample carrying a CALR
Type-1 mutation (52-bp deletion) and displaying a mutant allele burden
of approximately 50% according to sequencing analysis. Serial dilutions
were made up to 1% of the mutant in wild-type DNA. The CALR mutant could be detected in up to 2% dilution (Figure 1D).
Discussion
Recent methodological advances have shown that somatic mutations in the CALR gene occur in 20% to 25% of patients with ET and MF[10,11] and are now incorporated into the most recent revision of the WHO diagnostic criteria.[12] Determination of CALR mutations is essential for the diagnostic workup[12] and has an impact on the assessment of prognosis.[13-15]CALR mutation analysis is carried out with several methods including Sanger sequencing,[26] fragment analysis,[27] HRM-A,[19] Real Time PCR using TaqMan probes[28] and NGS[21]
with each one of these methods having their specific advantages and
drawbacks. Sanger sequencing and fragment analysis have limited
sensitivity (in the range of 10-25% and 5-10% respectively), while the
NGS technology presents the lowest LoD (1-1.5%) but is costly and
time-consuming for routine use, at present.[21,29]More
specifically, compared to NGS, our HRM-A protocol can be completed
within a few hours of sample collection, providing fast
turn-around times. On the other hand, NGS library preparation and
sequencing require a minimum of 2 days for completion, without taking
into account the downstream time-consuming analytical steps and the
prerequisite of multiplexing more than one sample for
cost-effectiveness. The latter is a major obstacle for labs with
small sample load. In addition, the NGS output is such that aiming
solely at the CALR mutation,
would be a waste of resources. To fully exploit the capabilities of
NGS, clinical laboratories either need a significant load of samples on
a routine basis or the simultaneous analysis of more than one genomic
region. For
all other occasions, HRM-A offers a relatively simple but equally
reliable technique that can support the daily routine of individual
samples with fast turn- around times.Other
platforms that offer equally satisfactory sensitivity levels to HRM-A
are TaqMan-based assays. The classical method uses fluorescently
labeled probes, that bind specific DNA sequences. In this way,
potentially unwanted PCR by-products can be ignored during analysis,
thus allowing for less stringent PCR conditions and primer design.
However, the use of nucleotide probes increases expenses and limits the
detection to previously determined mutations only. On the contrary, the
HRM-A approach is less costly and also permits the detection of novel
mutations. A potential drawback of the HRM-A method is the use of
fluorescent molecules that bind double-stranded nucleic acids in
a non-specific manner. This feature may lead to confusing detection
signals, due to potential secondary amplicons, unless experimental
settings are optimized in a way that allows the amplification of
specific products. This prerequisite has been fulfilled in our protocol
design.In this communication, we propose an optimized HRM-A protocol, which can identify CALR
exon 9 mutations on the basis of the differential plots that clearly
discriminate mutant from wild-type samples. Moreover, this version can
efficiently classify Type-1 and Type-2 common mutations and appears to
prevent the detection of false-positive results, in contrast to earlier
reports.[21] These characteristics are attributed to
the carefully designed highly specific primers, along with finely
tunned primer annealing temperature (Ta) and PCR cycling conditions.
Our modification also allowed prompt detection of novel mutations
through the appearance of variable changes in the melting curve profile
of undetermined genotype samples, which were then further investigated
using Sanger sequencing. In our experiments, the LoD of mutated DNA in
a wild- type background was 2%, which is considered fully acceptable
(≥2%), marginally lower than that previously reported when using the
same technique (3%) and not substantially different than the respective
TaqMan assays, where LoD varies between 1-3%.[19,28]
In
order to validate our standardized technique, we applied it in a study
of a large cohort of Greek ET and MF patients. We observed that neither
the overall frequency of the CALR
exon 9 mutations nor their distribution in the Greek ET and MF patients
was substantially different from those reported in similar surveys from
other European institutes.[10,11,30,31] In addition, we identified six novel mutations, which are to be added in the COSMIC database. Conclusions
The
proposed modification of the HRM- A technique is considered reliable
and has proven useful for the large-scale survey of the CALR exon 9 mutations across the Greek ET and MF patients.
References
- Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri
SA, Stein H, Thiele J, Vardiman JW, eds. WHO classification of tumours
of haematopoietic and lymphoid tissues, 4th Ed., Lyon, IARC. 2008,
40-50. available at: http://apps.who.int/bookorders/anglais/detart1.jsp?codlan=1&codcol= 70&codcch=4002 (Aug. 2018).
- Baxter
EJ, Scott LM, Campbell PJ, East C, Fourouclas N, Swanton S, Vassiliou
GS, Bench AJ, Boyd EM, Curtin N, Scott MA, Erber WN, Green AR; Cancer
Genome Project. Acquired mutation of the tyrosine kinase JAK2 in human
myeloproliferative disorders. Lancet 2005; 365(9464): 1054-1061.
available at: https://www.thelancet.com/journals/lancet/article/PIIS0140- 6736(05)71142-9/abstract (Aug. 2018).
- James
C, Ugo V, Le Couédic JP, Staerk J, Delhommeau F, Lacout C, Garçon L,
Raslova H, Berger R, Bennaceur-Griscelli A, Villeval JL, Constantinescu
SN, Casadevall N, Vainchenker W. A unique clonal JAK2 mutation leading
to constitutive signalling causes polycythaemia vera. Nature 2005;
434(7037): 1144-1148. available at: https://www.nature.com/articles/nature03546 (Aug. 2018).
- Jones
AV, Kreil S, Zoi K, Waghorn K, Curtis C, Zhang L, Score J, Seear R,
Chase AJ, Grand FH, White H, Zoi C, Loukopoulos D, Terpos E, Vervessou
EC, Schultheis B, Emig M, Ernst T, Lengfelder E, Hehlmann R, Hochhaus
A, Oscier D, Silver RT, Reiter A, Cross NC. Widespread occurrence of
the JAK2V617F mutation in chronic myeloproliferative disorders. Blood
2005; 106(6): 2162-2168. available at: http://www.bloodjournal.org/content/106/6/2162.long (Aug. 2018).
- Kralovics
R, Passamonti F, Buser AS, Teo SS, Tiedt R, Passweg JR, Tichelli A,
Cazzola M, Skoda RC. A gain-of-function mutation of JAK2 in
myeloproliferative disorders. N Engl J Med 2005; 352(17): 1779-1790.
available at: https://www.nejm.org/doi/full/10.1056/NEJMoa051113 (Aug. 2018).
- Levine
RL, Loriaux M, Huntly BJ, Loh ML, Beran M, Stoffregen E, Berger R,
Clark JJ, Willis SG, Nguyen KT, Flores NJ, Estey E, Gattermann N,
Armstrong S, Look AT, Griffin JD, Bernard OA, Heinrich MC, Gilliland
DG, Druker B, Deininger MW. The JAK2V617F activating mutation occurs in
chronic myelomonocytic leukemia and acute myeloid leukemia, but not in
acute lymphoblastic leukemia or chronic lymphocytic leukemia. Blood
2005; 106(10): 3377-3379. available at: http://www.bloodjournal.org/content/106/10/3377/tab-figures-only (Aug. 2018).
- Scott
LM, Tong W, Levine RL, Scott MA, Beer PA, Stratton MR, Futreal PA,
Erber WN, McMullin MF, Harrison CN, Warren AJ, Gilliland DG, Lodish HF,
Green AR. JAK2 exon 12 mutations in polycythemia vera and idiopathic
erythrocytosis. N Engl J Med 2007; 356(5): 459-468. available at: https://www.nejm.org/doi/full/10.1056/NEJMoa065202 (Aug. 2018).
- Pikman
Y, Lee BH, Mercher T, McDowell E, Ebert BL, Gozo M, Cuker A, Wernig G,
Moore S, Galinsky I, DeAngelo DJ, Clark JJ, Lee SJ, Golub TR, Wadleigh
M, Gilliland DG, Levine RL. MPLW515L is a novel somatic activating
mutation in myelofibrosis with myeloid metaplasia. PLoS Med 2006; 3(7):
e270. available at: https://journals.plos.org/plosmedicine/article?id=10.1371/journal.pme d.0030270 (Aug. 2018).
- Vardiman
JW, Thiele J, Arber DA, Brunning RD, Borowitz MJ, Porwit A, Harris NL,
Le Beau MM, Hellström-Lindberg E, Tefferi A, Bloomfield CD. The 2008
revision of the World Health Organization (WHO) classification of
myeloid neoplasms and acute leukemia: rationale and important changes.
Blood 2009; 114(5): 937-951. available at: http://www.bloodjournal.org/content/early/2009/04/08/blood-2009- 03-209262 (Aug. 2018).
- Klampfl
T, Gisslinger H, Harutyunyan AS, Nivarthi H, Rumi E, Milosevic JD, Them
NC, Berg T, Gisslinger B, Pietra D, Chen D, Vladimer GI, Bagienski K,
Milanesi C, Casetti IC, Sant'Antonio E, Ferretti V, Elena C, Schischlik
F, Cleary C, Six M, Schalling M, Schönegger A, Bock C, Malcovati L,
Pascutto C, Superti-Furga G, Cazzola M, Kralovics R. Somatic mutations
of calreticulin in myeloproliferative neoplasms. N Engl J Med 2013;
369(25): 2379- 2390. available at: https://www.nejm.org/doi/full/10.1056/NEJMoa1311347 (Aug. 2018).
- Nangalia
J, Massie CE, Baxter EJ, Nice FL, Gundem G, Wedge DC, Avezov E, Li J,
Kollmann K, Kent DG, Aziz A, Godfrey AL, Hinton J, Martincorena I, Van
Loo P, Jones AV, Guglielmelli P, Tarpey P, Harding HP, Fitzpatrick JD,
Goudie CT, Ortmann CA, Loughran SJ, Raine K, Jones DR, Butler AP,
Teague JW, O'Meara S, McLaren S, Bianchi M, Silber Y, Dimitropoulou D,
Bloxham D, Mudie L, Maddison M, Robinson B, Keohane C, Maclean C, Hill
K, Orchard K, Tauro S, Du MQ, Greaves M, Bowen D, Huntly BJP, Harrison
CN, Cross NCP, Ron D, Vannucchi AM, Papaemmanuil E, Campbell PJ, Green
AR. Somatic CALR mutations in myeloproliferative neoplasms with
nonmutated JAK2. N Engl J Med 2013; 369(25): 2391-2405. available at: https://www.nejm.org/doi/full/10.1056/NEJMoa1312542 (Aug. 2018).
- Arber
DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM,
Bloomfield CD, Cazzola M, Vardiman JW. The 2016 revision to the World
Health Organization classification of myeloid neoplasms and acute
leukemia. Blood 2016; 127(20): 2391-2405. available at: http://www.bloodjournal.org/content/early/2016/04/11/blood-2016- 03-643544 (Aug. 2018).
- Rotunno
G, Mannarelli C, Guglielmelli P, Pacilli A, Pancrazzi A, Pieri L,
Fanelli T, Bosi A, Vannucchi AM; Associazione Italiana per la Ricerca
sul Cancro Gruppo Italiano Malattie Mieloproliferative Investigators.
Impact of calreticulin mutations on clinical and hematological
phenotype and outcome in essential thrombocythemia. Blood 2014;
123(10): 1552-1555. available at: http://www.bloodjournal.org/content/123/10/1552 (Aug. 2018).
- Rumi
E, Pietra D, Ferretti V, Klampfl T, Harutyunyan AS, Milosevic JD, Them
NC, Berg T, Elena C, Casetti IC, Milanesi C, Sant'antonio E, Bellini M,
Fugazza E, Renna MC, Boveri E, Astori C, Pascutto C, Kralovics R,
Cazzola M; Associazione Italiana per la Ricerca sul Cancro Gruppo
Italiano Malattie Mieloproliferative Investigators. JAK2 or CALR
mutation status defines subtypes of essential thrombocythemia with
substantially different clinical course and outcomes. Blood 2014;
123(10): 1544-1551. available at: http://www.bloodjournal.org/content/123/10/1544?sso-checked=true (Aug. 2018).
- Tefferi
A, Lasho TL, Finke CM, Knudson RA, Ketterling R, Hanson CH, Maffioli M,
Caramazza D, Passamonti F, Pardanani A. CALR vs JAK2 vs MPL-mutated or
triple-negative myelofibrosis: clinical, cytogenetic and molecular
comparisons. Leukemia 2014; 28(7): 1472- 1477. available at: https://www.nature.com/articles/leu20143 (Aug. 2018).
- Tefferi
A, Guglielmelli P, Lasho TL, Rotunno G, Finke C, Mannarelli C, Belachew
AA, Pancrazzi A, Wassie EA, Ketterling RP, Hanson CA, Pardanani A,
Vannucchi AM. CALR and ASXL1 mutations- based molecular prognostication
in primary myelofibrosis: an international study of 570 patients.
Leukemia 2014; 28(7): 1494-1500. available at: https://www.nature.com/articles/leu201457 (Aug. 2018).
- Nangalia
J, Green TR. The evolving genomic landscape of myeloproliferative
neoplasms. Hematology Am Soc Hematol Educ Program 2014; 2014(1):
287-296. available at: http://asheducationbook.hematologylibrary.org/content/2014/1/287.lo ng (Aug. 2018).
- Bench
AJ, White HE, Foroni L, Godfrey AL, Gerrard G, Akiki S, Awan A, Carter
I, Goday-Fernandez A, Langabeer SE, Clench T, Clark J, Evans PA,
Grimwade D, Schuh A, McMullin MF, Green AR, Harrison CN, Cross NC;
British Committee for Standards in Haematology. Molecular diagnosis of
the myeloproliferative neoplasms: UK guidelines for the detection of
JAK2V617F and other relevant mutations. Br J Haematol 2013; 160(1):
25-34. available at: https://onlinelibrary.wiley.com/doi/abs/10.1111/bjh.12075 (Aug. 2018).
- Bilbao-Sieyro
C, Santana G, Moreno M, Torres L, Santana-Lopez G, Rodriguez-Medina C,
Perera M, Bellosillo B, de la Iglesia S, Molero T, Gomez-Casares MT.
High resolution melting analysis: a rapid and accurate method to detect
CALR mutations. PLoS One 2014; 9(7): e103511. available at: https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0103 511 (Aug. 2018).
- Chi
J, Nicolaou KA, Nicolaidou V, Koumas L, Mitsidou A, Pierides C,
Manoloukos M, Barbouti K, Melanthiou F, Prokopiou C, Vassiliou GS
and Costeas P. Calreticulin gene exon 9 frameshift mutations in
patients with thrombocytosis. Leukemia 2014; 28(5): 1152-1154.
available at: https://www.nature.com/articles/leu2013382 (Aug. 2018).
- Jones
AV, Ward D, Lyon M, Leung W, Callaway A, Chase A, Dent CL, White HE,
Drexler HG, Nangalia J, Mattocks C, Cross NC. Evaluation of methods to
detect CALR mutations in myeloproliferative neoplasms. Leuk Res 2015;
39(1): 82-87. available at: https://www.lrjournal.com/article/S0145-2126(14)00371-3/abstract (Aug. 2018).
- Vannucchi
AM, Rotunno G, Bartalucci N, Raugei G, Carrai V, Balliu M, Mannarelli
C, Pacilli A, Calabresi L, Fjerza R, Pieri L, Bosi A, Manfredini R,
Guglielmelli P. Calreticulin mutation-specific immunostaining in
myeloproliferative neoplasms: pathogenetic insight and diagnostic
value. Leukemia 2014; 28(9): 1811-1818. available at: https://www.nature.com/articles/leu2014100 (Aug. 2018).
- Vossen
RH, Aten E, Roos A, den Dunnen JT. High-resolution melting analysis
(HRMA): more than just sequence variant screening. Hum Mutat 2009;
30(6): 860-866. available at: http://www.bloodjournal.org/content/111/8/4418 (Aug. 2018).
- Bergamaschi
GM, Primignani M, Barosi G, Fabris FM, Villani L, Reati R, Dell'era A,
Mannucci PM. MPL and JAK2 exon 12 mutations in patients with the
Budd-Chiari syndrome or extrahepatic portal vein obstruction. Blood
2008; 111(8): 4418.
- Forbes SA, Beare D,
Boutselakis H, Bamford S, Bindal N, Tate J, Cole CG, Ward S, Dawson E,
Ponting L, Stefancsik R, Harsha B, Kok CY, Jia M, Jubb H, Sondka Z,
Thompson S, De T, Campbell PJ. COSMIC: somatic cancer genetics at
high-resolution. Nucleic Acids Res 2017; 45(Database issue): D777–D783.
available at: https://academic.oup.com/nar/article/45/D1/D777/2605743?searchres ult=1 (Aug. 2018).
- Mehrotra
M, Luthra R, Singh RR, Barkoh BA, Galbincea J, Mehta P, Goswami RS,
Jabbar KJ, Loghavi S, Medeiros LJ, Verstovsek S, Patel KP. Clinical
validation of a multipurpose assay for detection and genotyping of CALR
mutations in myeloproliferative neoplasms. Am J Clin Pathol 2015;
144(5): 746-755. available at: https://academic.oup.com/ajcp/article/144/5/746/1761207?searchresul t=1 (Aug. 2018).
- Maier
CL, Fisher KE, Jones HH, Hill CE, Mann KP, Zhang L. Development and
validation of CALR mutation testing for clinical diagnosis. Am J Clin
Pathol 2015; 144(5): 738-745. available at: https://academic.oup.com/ajcp/article/144/5/738/1761139?searchresul t=1 (Aug. 2018).
- Chi
J, Manoloukos M, Pierides C, Nicolaidou V, Nicolaou K, Kleopa M,
Vassiliou G, Costeas P. Calreticulin mutations in myeloproliferative
neoplasms and new methodology for their detection and monitoring. Ann
Hematol 2015; 94(3): 399-408. available at: https://link.springer.com/article/10.1007/s00277-014- 2232-8 (Aug. 2018).
- Luo
W, Zhongxin Yu Z. Calreticulin (CALR) mutation in myeloproliferative
neoplasms (MPNs). Stem Cell Investig 2015; 2: 16. available at: http://sci.amegroups.com/article/view/7264/8051 (Aug. 2018).
- Rumi
E, Pietra D, Pascutto C, Guglielmelli P, Martínez-Trillos A, Casetti I,
Colomer D, Pieri L, Pratcorona M, Rotunno G, Sant'Antonio E, Bellini M,
Cavalloni C, Mannarelli C, Milanesi C, Boveri E, Ferretti V, Astori C,
Rosti V, Cervantes F, Barosi G, Vannucchi AM, Cazzola M; Associazione
Italiana per la Ricerca sul Cancro Gruppo Italiano Malattie
Mieloproliferative Investigators. Clinical effect of driver mutations
of JAK2, CALR, or MPL in primary myelofibrosis. Blood. 2014; 124(7):
1062-1069. available at: http://www.bloodjournal.org/content/124/7/1062 (Aug. 2018).
- Cabagnols
X, Defour JP, Ugo V, Ianotto JC, Mossuz P, Mondet J, Girodon F,
Alexandre JH, Mansier O, Viallard JF, Lippert E, Murati A, Mozziconacci
MJ, Saussoy P, Vekemans MC, Knoops L, Pasquier F, Ribrag V, Solary E,
Plo I, Constantinescu SN, Casadevall N, Vainchenker W, Marzac C,
Bluteau O. Differential association of calreticulin type 1 and type 2
mutations with myelofibrosis and essential thrombocytemia: relevance
for disease evolution. Leukemia. 2015; 29(1): 249-252. available at: http://www.bloodjournal.org/content/124/21/1823?sso-checked=true (Aug. 2018).
[TOP]


