Yi-Chang Liu1,2, Ching-Ping Lee1, Tsung-Jang Yeh1, Yuh-Ching Gau1, Chieh-Yu Hsieh1, Ya-Lun Ke1, Jeng-Shiun Du1, Ming-Hui Lin1, Hui-Ching Wang1, Shih-Hao Tang1, Shih-Feng Cho1,2, Chi-En Hsiao1, Jui-Feng Hsu1, Samuel Yien Hsiao4, Chin-Mu Hsu1,3 and Hui-Hua Hsiao1,2,3.
1 Division
of Hematology and Oncology, Department of Internal Medicine, Kaohsiung
Medical University Hospital, Kaohsiung, Taiwan.
2 Faculty of Medicine, Kaohsiung Medical University, Kaohsiung, Taiwan.
3 Center for Liquid Biopsy, Kaohsiung Medical University, Kaohsiung, Taiwan.
4 University of Rutgers-Camden, New Jersey, USA.
Corresponding
author: Dr. Hui-Hua Hsiao. Address: 100, Tzu-You 1st road, San-Ming
Dist., Kaohsiung, Taiwan. Tel: +886-7-3121101#6110; Fax:
+886-7-3162429. E-mail:
huhuhs@kmu.edu.tw
Published: May 1, 2020
Received: December 27, 2020
Accepted: April 2, 2020
Mediterr J Hematol Infect Dis 2020, 12(1): e2020022 DOI
10.4084/MJHID.2020.022
This is an Open Access article distributed
under the terms of the Creative Commons Attribution License
(https://creativecommons.org/licenses/by-nc/4.0),
which permits unrestricted use, distribution, and reproduction in any
medium, provided the original work is properly cited.
|
To the Editor,
Myeloproliferative
neoplasms (MPN), characterized by abnormal proliferation of myeloid
series, were found to have specific molecular markers.[1-5]
In Philadelphia-negative and BCR-ABL-negative patients, JAK2 V617F
mutation is the major marker and is present in approximately 90% in
polycythemia vera patients and about 50~60% of patients with essential
thrombocythemia (ET) and primary myelofibrosis (MF) patients.[1-4] The identification of this mutation contributes to the diagnosis and prognostic significance, as reported in many articles.[1-4]
More recently, several investigations identified novel somatic
mutations at the exon 9 of calreticulin (CALR) gene in 50~80% of
JAK2V617F-negative ET and MF patients.[5]
CALR is
a highly conserved endoplasmic reticulum calcium-binding chaperone that
is related to calcium homeostasis, cell adhesion, and immune response.[9]
Until now, all CALR mutations have been found at exon 9, which encodes
the C-domain region where there is a domain for Ca2+ to bind.[5-9,14,15]
Of the more than 50 mutations found, all were exclusively on JAK2
V617F-negative patients. Most of the mutations correspond to a 52 kb
deletion (del 1092-1143, type 1) or a 5bp insertion (1151 ins TTGTC,
type 2), resulting in premature terminations from these frameshift
mutations.[7,8] From previous studies, the CALR
mutations had a unique clinical presentation compared with JAK2 V617F
mutation; therefore, it is essential to survey these mutations in
Philadelphia chromosome and/or BCR-ABL negative MPNs.[10-14]
High
resolution melting (HRM) method using the saturating dsDNA binding dye
for melting curve analysis is a rapid and labor-saving method for
mutation screening.[15] In this study, we used the
HRM method to screen for these mutations in our ET patients by
distinguishing the specific curve types. We also reviewed the clinical
presentations of patients to find the relationship between mutations
and clinical phenotypes.
Materials and Methods
Patients and samples.
A total of 60 consecutive patients diagnosed with ET, according to
Polycythemia vera Study Group criteria at Kaohsiung Medical University
Chung-Ho Memorial Hospital, were enrolled in the study with informed
consent. The DNA from whole peripheral blood was isolated using the
QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany), according to the
manufacturer’s protocol. The quality and concentration of DNA were
determined by NanoVue Plus Spectrophotometer (GE Healthcare, UK) and
stored at -20 °C for further use.
JAK2 V617F mutation survey. The JAK2 V617F mutation was identified by amplification refractory mutation system (ARMS).[4]
CALR mutation survey.
For the HRM survey, the reaction was performed by the ABI ViiA7
machine. Briefly, 20 ng of DNA and 5 μM primers were amplified with the
final volume of 20 μl with HRM reaction kit (MeltDoctor HRM Master Mix,
Thermo fisher scientific, Waltham, MA, USA). The primer sets were
designed (Primer Express® Software 3.0, Thermo fisher scientific,
Waltham, MA, USA) with forward: 5’-GGCCTCTCTACAGCTCGTCCTT-3’ and
reverse 5’-ACGTCCGTCGTCTCTTTGTT-3’. The amplification conditions are as
follows: an initial denaturation step at 95 °C for 10 min, followed by
40 cycles of denaturation at 95 °C for 15 s and annealing/extension at
62 °C for 1 min, with the acquisition of fluorescent signals at the end
of each extension step. The fluorescent signals for HRM analysis were
detected at 0.2 °C intervals, with hold-time for 10 s, between 95-60
°C. The selected primer set amplified 213 bp amplicon, and results were
analyzed as fluorescence versus temperature graphs by software with
normalized, temperature-shifting melting cures display as a difference
plot.
For sequencing analysis, the DNA samples were carried out
polymerase chain reaction (PCR) used with primer sets of forward:
5’-ACAACTTCCTCATCACCAACG-3’ and reverse: 5’-GGCCTCAGTCCAGCCCTG-3’. The
amplicons were performed with BigDye Terminator v3.1 Cycle Sequencing
Kit (Thermo Fisher Scientific, Waltham, MA, USA) by bidirectional
sequencing and analyzed by ABI 3730 XL DNA sequencing analyzer (Applied
Biosystems, CA, USA). The sequencing results were compared to reference
sequences of the CALR gene available on GenBank (www.ncbi.nlm.nih.gov/genbank/) to verify the difference.
Statistical Analysis.
Statistical analyses were performed by SPSS software ver.19 for Windows
(SPSS Inc., Chicago, IL, USA). The chi-square test was used to test
categorical variables, while the Student’s t-test compared continuous
variables. A p-value of < 0.05 considered statistically significant.
Results
Of
the ET patients studied, 96.6% have somatic mutations with JAK2 V617F
and CALR mutations. Specifically, of the 60 ET patients studied, 34
(56.7%) were JAK2 V617F positive. Of the 26 JAK2V617F negative
patients, 21 (80.8%) had CALR mutations were detected by HRM
analysis with eight types of melting curves (Figure 1).
These CALR mutations, confirmed by direct sequencing method, spread
over exon 9, including del 1092-1143 mutations (type 1, 6 persons;
28.5%), 1151 ins TTGTC mutations (type 2, 7 patients; 33.3%), 3
complicated mutations and 3 other mutations (Table 1).
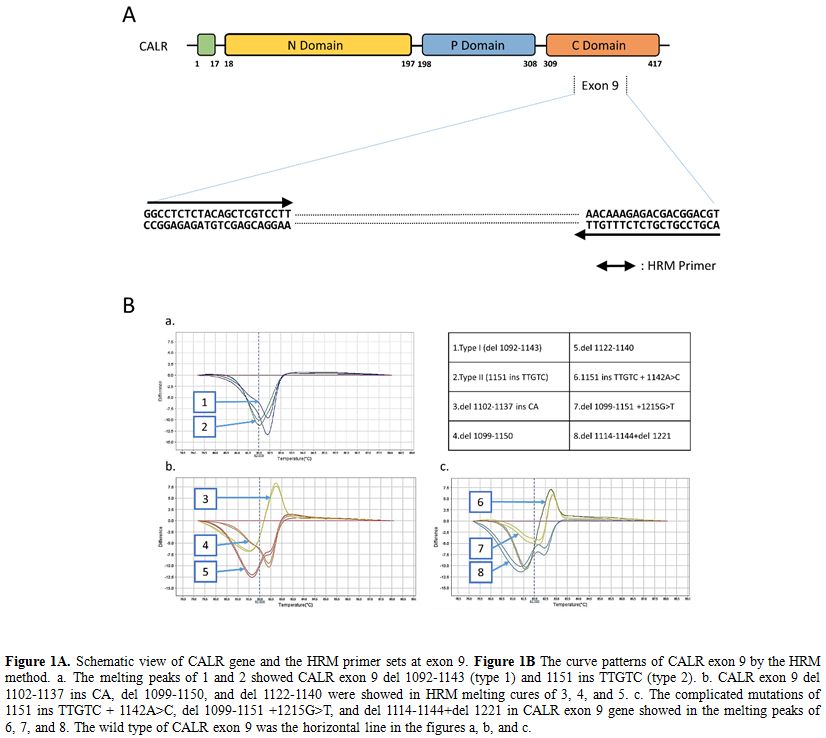 |
Figure 1. Figure
1A. Schematic view of CALR gene and the HRM primer sets at exon 9. Figure 1B
The curve patterns of CALR exon 9 by the HRM method. a. The melting
peaks of 1 and 2 showed CALR exon 9 del 1092-1143 (type 1) and 1151 ins
TTGTC (type 2). b. CALR exon 9 del 1102-1137 ins CA, del 1099-1150, and
del 1122-1140 were showed in HRM melting cures of 3, 4, and 5. c. The
complicated mutations of 1151 ins TTGTC + 1142A>C, del 1099-1151
+1215G>T, and del 1114-1144+del 1221 in CALR exon 9 gene showed in
the melting peaks of 6, 7, and 8. The wild type of CALR exon 9 was the
horizontal line in the figures a, b, and c. |
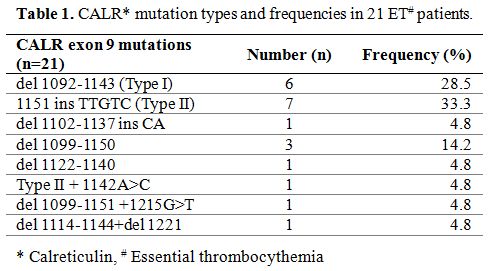 |
Table 1. CALR* mutation types and frequencies in 21 ET# patients. |
The
clinical presentation revealed that there was no significant difference
over gender between JAK2 and CALR mutations. However, patients with
CALR mutations were younger and had a higher platelet count than
patients with JAK2 mutation significantly (p<0.05) (Table 2).
Patients with JAK2 mutations had significantly higher leukocytes and
hemoglobin levels and more thrombotic events than patients with CALR
mutations (p<0.05).
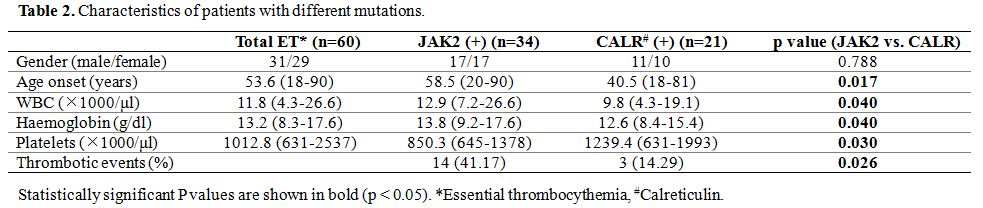 |
Table 2. Characteristics of patients with different mutations. |
Discussion
Besides
JAK2V617F mutation, recent studies showed that somatic mutation at exon
9 of CALR accounts of 50~80% JAK2V617F-negative ET and MF patients.[5-9]
It has diagnostic importance and also contributes to the clinical
characteristics of ET patients; therefore, it is crucial to identify
the mutations.[10-15] Although most of the mutations
correspond to a 52 bp deletion (type 1) or a 5 bp insertion (type 2),
there were more than 50 other mutations found. The variations of
mutations continue to pose a challenge for a rapid and effective
survey.[15]
The incidence of CALR mutation in
our patients was similar to previous reports. However, the frequency of
type 1 mutation was lower, with only half of the mutated patients
presenting either type 1 or 2 mutations. The distribution was different
from other reports.[7,8,10,13,14,15] Recently, Keaney and Li et al., reported a lower incidence of type 1 mutation in myelofibrosis patients.[10,12]
These data might suggest that there is a population difference in CALR
mutations and highlight the need for further mutation survey outside
type 1 and 2.
HRM system using the saturating dsDNA binding dye
for mutation survey and genotyping without the need for costly labeled
oligonucleotides is an important step forward for mutation survey. In
previous studies with HRM methods, it could identify type 1 and 2
mutations effectively and rapidly[13,15]
were also shown in our studies. Our results were able to detect 6 other
mutation types concurrently and clearly by the HRM method relating to
the CALR mutations even when the mutation is outside type 1 or 2.
Our
previous reports showed typical presentations of JAK2 V617F mutation
patients, characterized by frequent leukocytosis and thrombosis,[2] here, the clinical presentations of the two groups with either JAK2 or CALR mutation were similar to previous reports.[5,11,14]
Patients with JAK2 mutations had a higher WBC count, hemoglobin level,
and thrombotic events, while patients with CALR mutations were younger
and had a significantly higher platelet count. Some reports also
demonstrated different clinical manifestations between type 1 and 2
CALR mutations;[10,14-15] however, due to the small number of studied cases, no differences were shown in this study.
Conclusions
The
HRM method provides a fast and effective tool for the identification of
clinical variants of CALR genes and the understanding of the
predisposition of the disease. This report is the first on ET cases in
Taiwanese cases by HRM method, and we identified six mutations that
were outside type 1 or 2 mutations on CALR that may indicate a
predisposition to the disease. Patients with CALR mutation had
different clinical presentations when compared to JAK2 mutation and
should be checked.
References
- Levine RL, Wadleigh M, Cools J, et al. Activating
mutation in the tyrosine kinase JAK2 in polycythemia vera, essential
thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer
Cell. 2005;7:387-397. https://doi.org/10.1016/j.ccr.2005.03.023 PMid:15837627
- Hsiao
HH, Yang MY, Liu YC, Lee CP, Yang WC, Liu TC, Chang CS, Lin SF. The
association of JAK2V617F mutation and leukocytosis with thrombotic
events in essential thrombocythemia. Exp Hematol. 2007;35:1704-1707. https://doi.org/10.1016/j.exphem.2007.08.011 PMid:17920754
- Spanoudakis
E, Papoutselis M, Bazdiara I, Lamprianidi E, Kordella X, Tilkeridis C,
Tsatalas C, Kotsianidis I. The JAK2V617F Point Mutation Increases the
Osteoclast Forming Ability of Monocytes in Patients with Chronic
Myeloproliferative Neoplasms and Makes their Osteoclasts more
Susceptible to JAK2 Inhibition. Mediterr J Hematol Infect Dis.
2018;10:e2018058. https://doi.org/10.4084/mjhid.2018.058 PMid:30416690 PMCid:PMC6223546
- Leszczynska
A, Grzenkowicz-Wydra J, Chmielewska-Gorycka L, Bieniaszewska M,
Hellmann A. Detection of JAK2 Exon 12 Mutations in JAK2 V617F-Negative
Polycythemia Vera Patients by Cloning Technique. Acta Haematol.
2016;136:123-128. https://doi.org/10.1159/000446798 PMid:27410038
- Nangalia
J, Massie CE, Baxter EJ, Nice FL, Gundem G, Wedge DC, Avezov E, Li J,
Kollmann K, Kent DG, Aziz A, Godfrey AL, Hinton J, Martincorena I, Van
Loo P, Jones AV, Guglielmelli P, Tarpey P, Harding HP, Fitzpatrick JD,
Goudie CT, Ortmann CA, Loughran SJ, Raine K, Jones DR, Butler AP,
Teague JW, O'Meara S, McLaren S, Bianchi M, Silber Y, Dimitropoulou D,
Bloxham D, Mudie L, Maddison M, Robinson B, Keohane C, Maclean C, Hill
K, Orchard K, Tauro S, Du MQ, Greaves M, Bowen D, Huntly BJP, Harrison
CN, Cross NCP, Ron D, Vannucchi AM, Papaemmanuil E, Campbell PJ, Green
AR. Somatic CALR mutations in myeloproliferative neoplasms with
nonmutated JAK2. N Engl J Med. 2013;369:2391-2405. https://doi.org/10.1056/NEJMoa1312542 PMid:24325359 PMCid:PMC3966280
- Clinton A, McMullin MF. The Calreticulin gene and myeloproliferative neoplasms. J Clin Pathol. 2016;69:841-845. https://doi.org/10.1136/jclinpath-2016-203899 PMid:27354406
- Wong
WJ, Hasserjian RP, Pinkus GS, Breyfogle LJ, Mullally A, Pozdnyakova O.
JAK2, CALR, MPL and ASXL1 mutational status correlates with distinct
histological features in Philadelphia chromosome-negative
myeloproliferative neoplasms. Haematologica. 2018;103:e63-e68. https://doi.org/10.3324/haematol.2017.178988 PMid:29146710 PMCid:PMC5792288
- Xia
D, Hasserjian RP. Molecular testing for JAK2, MPL, and CALR in
myeloproliferative neoplasms. Am J Hematol. 2016;91:1277-1280. https://doi.org/10.1002/ajh.24578 PMid:27727468
- Misawa
K, Yasuda H, Araki M, Ochiai T, Morishita S, Shirane S, Edahiro Y,
Gotoh A, Ohsaka A, Komatsu N. Mutational subtypes of JAK2 and CALR
correlate with different clinical features in Japanese patients with
myeloproliferative neoplasms. Int J Hematol. 2018;107:673-680. https://doi.org/10.1007/s12185-018-2421-7 PMid:29464483
- Keaney
T, O'Connor L, Krawczyk J, Abdelrahman MA, Hayat AH, Murray M, O'Dwyer
M, Percy M, Langabeer S, Haslam K, Glynn B, Mullen C, Kead Lahiff S,
Smith TJ. A novel molecular assay using hybridisation probes and melt
curve analysis for CALR exon 9 mutation detection in myeloproliferative
neoplasms. J Clin Pathol. 2017;70:662-668. https://doi.org/10.1136/jclinpath-2016-204205 PMid:28143941
- Didone
A, Nardinelli L, Marchiani M, Ruiz ARL, de Lima Costa AL, Lima IS,
Santos NM, Sanabani SS, Bendit I. Comparative study of different
methodologies to detect the JAK2 V617F mutation in chronic BCR-ABL1
negative myeloproliferative neoplasms. Pract Lab Med. 2016;4:30-37. https://doi.org/10.1016/j.plabm.2015.12.004 PMid:28856190 PMCid:PMC5574508
- Li
B, Xu J, Wang J, Gale RP, Xu Z, Cui Y, Yang L, Xing R, Ai X, Qin T,
Zhang Y, Zhang P, Xiao Z. Calreticulin mutations in Chinese with
primary myelofibrosis. Haematologica. 2014;99:1697-1700. https://doi.org/10.3324/haematol.2014.109249 PMid:24997152 PMCid:PMC4222480
- Pavlov
I, Hadjiev E, Alaikov T, Spassova S, Stoimenov A, Naumova E, Shivarov
V, Ivanova M. Calreticulin Mutations in Bulgarian MPN Patients. Pathol
Oncol Res. 2018;24:171-174. https://doi.org/10.1007/s12253-017-0226-2 PMid:28411309
- Zini
R, Guglielmelli P, Pietra D, Rumi E, Rossi C, Rontauroli S, Genovese E,
Fanelli T, Calabresi L, Bianchi E, Salati S, Cazzola M, Tagliafico E,
Vannucchi AM, Manfredini R, investigators A. CALR mutational status
identifies different disease subtypes of essential thrombocythemia
showing distinct expression profiles. Blood Cancer J. 2017;7:638 https://doi.org/10.1038/s41408-017-0010-2 PMid:29217833 PMCid:PMC5802509
- Giannopoulos
A, Rougkala N, Loupis T, Mantzourani M, Viniou NA, Variami E,
Vassilakopoulos TP, Dryllis G, Kotsianidis I, Gougopoulou T, Politou M,
Konstantopoulos K, Vassilopoulos G. Detection of CALR Mutations Using
High Resolution Melting Curve Analysis (HRM-A); Application on a Large
Cohort of Greek ET and MF patients. Mediterr J Hematol Infect Dis.
2019;11:e2019009. https://doi.org/10.4084/mjhid.2019.009 PMid:30671215 PMCid:PMC6328041
[TOP]


