Sumalai Dechyotin1, Kittipong Sakunthai1, Noppmats Khemtonglang2, Supawadee Yamsri2,3, Kanokwan Sanchaisuriya2,3, Kriengkrai Kitcharoen2 and Suttiphan Kitcharoen2.
1 Medical Science Program, Graduate School, Khon Kaen University, Thailand.
2 Department of Medical Technology, Faculty of Associated Medical Sciences, Khon Kaen University, Thailand.
3
Center for Research and Development of Medical Diagnostic Laboratories,
Faculty of Associated Medical Sciences, Khon Kaen University, Thailand.
Correspondence to: Dr.
Suttiphan Kitcharoen. Department of Medical Technology, Faculty of
Associated Medical Sciences, Khon Kaen University, Khon Kaen 40002,
Thailand. E-mail:
sutpra@kku.ac.th
Published: May 1, 2021
Received: November 04, 2020
Accepted: April 6, 2021
Mediterr J Hematol Infect Dis 2021, 13(1): e2021029 DOI
10.4084/MJHID.2021.029
This is an Open Access article distributed
under the terms of the Creative Commons Attribution License
(https://creativecommons.org/licenses/by-nc/4.0),
which permits unrestricted use, distribution, and reproduction in any
medium, provided the original work is properly cited.
|
|
Abstract
Introduction:
Glucose-6-phosphate dehydrogenase (G6PD) deficiency is the most common
X-linked enzymopathy, highly prevalent in the areas where malaria is or
has been endemic. Prevalence of G6PD deficiency and characterization of
G6PD variants in females from previously malaria-endemic areas of
northeastern Thailand remain unstudied.
Methods:
Prevalence of G6PD deficiency was determined by a fluorescent spot test
(FST), quantitative G6PD activity assay, and multiplex allele-specific
(AS)- and restriction fragment length polymorphic (RFLP)-PCR developed
for detection of common G6PD variants in the Thai population.
Results:
Prevalence of G6PD deficiency in female samples (n = 355) was 18% by
FST, 29.6% by quantitation of G6PD activity, and 28.1% by PCR-based
genotyping. The most common variant was G6PD Viangchan (54%), followed
by G6PD Canton (11%) and G6PD Union (11%); in addition, a novel
heterozygous variant, G6PD Khon Kaen (c.305T>C, p.F102S), was
identified. The majority of heterozygotes expressed G6PD activity
within the intermediate deficiency range (30-70% median of normal
enzyme activity).
Conclusion:
High prevalence of G6PD deficiency was present in females from
northeastern Thailand, the majority being due to heterozygosity of G6PD
variants. The findings will have a bearing on the inclusion of
primaquine in antimalarial-based policies for malaria elimination in
populations with a high prevalence of G6PD deficiency.
|
Introduction
Glucose-6-phosphate
dehydrogenase (G6PD) deficiency is the most common X-linked genetic
enzyme disorder. It is recently estimated to affect over 500 million
individuals worldwide.[1] G6PD deficiency arises from
mutations in G6PD resulting in low levels of enzyme activity, giving
rise to a spectrum of disorders, ranging from neonatal
hyperbilirubinemia to chronic non-spherocytic hemolytic anemia and
acute hemolysis. However, most people with this condition are
asymptomatic but can undergo hemolytic crisis when exposed to
infection, certain types of food (e.g., fava beans), or oxidant drugs
(e.g., 8-aminoquinoline antimalarials, primaquine, and tafenoquine).[1]
The World Health Organization (WHO) recommends neonatal screening for
G6PD deficiency in populations with a 3-5% prevalence of G6PD
deficiency.[2] In malaria-endemic countries, including
Thailand, the G6PD test is required before administering
8-aminoquinolines for the radical cure of Plasmodium vivax malaria to
eliminate latent stages in the liver (hypnozoites).[3,4]
G6PD
deficiency can be identified either by red blood cell (RBC) enzyme
activity assay (qualitative or quantitative) or genotyping of variants.[5] Quantitative
G6PD activity measurement (<30% normal activity) is sufficient to
identify male G6PD deficient hemizygotes and female homozygotes;[3,5]
however, for heterozygous females, a recent meta-analysis showed
inter-study variations in enzyme assay at the 70% level precludes a
universal diagnostic cutoff for G6PD deficiency.[6]Female heterozygotes are also at risk of neonatal hyperbilirubinemia[7,8] and vulnerable to hemolysis triggered by 8-aminoquinoline antimalarials.[9,10]
However, the severity of hemolysis depends on the dose of drugs and
phenotypes stemming from G6PD variants as the diversity of G6PD
variants leads to variations in residual enzyme activity and clinical
severity.[11,12] In heterozygous females, there
is substantial variability in the ratio of G6PD normal to
G6PD-deficient RBCs generated by Lyonization, and thus quantitation of
G6PD activity is superior to genotyping in predicting drug-induced
hemolysis,[1,9,11]Northeastern
Thailand shares borders with PDR Laos and Cambodia. Although these
regions were previously endemic for malaria, there is a looming risk of
reemergence of this infection, particularly now that P. falciparum
strains with reduced sensitivity to artemisinin, a first-line
antimalarial, are prevalent in the Greater Mekong Subregion (GMS).[13,14] Moreover, the predominant malaria parasite in the GMS tends to shift from P. falciparum to P. vivax,[15]
leading to an increased requirement for 8-aminoquinolines radical
treatment. The prevalence of G6PD deficiency in northeastern Thailand
has been reported to range from 8-24%.[16] However,
there are a limited number of reports on the prevalence of G6PD
deficiency in females in this region of the country; the majority of
studies are focused on the prevalence of G6PD deficiency in
hemizygotes.[16-20] A survey conducted in Burirum
Provincial Hospital on G6PD deficiency prevalence using a quantitative
G6PD activity assay of cord blood samples from male and female newborns
reported prevalence of G6PD deficiency of 21.7 and 8%, respectively.[21]Here,
the prevalence of G6PD deficiency in women of reproductive age residing
in northeastern Thailand was carried out by a fluorescent spot test
(FST), measurement of G6PD activity, and genotyping of G6PD variants.
The findings should contribute to determining the relationship between
G6PD variants and enzyme deficiencies in this region of the country. Materials and Methods
Study population.
Women of reproductive age (n = 355), 18-45 years of age, from
northeastern Thailand who initially enrolled in a screening program for
anemia between January and May 2017 were recruited.[22] All participants appeared physically healthy. The
research protocol was approved by the Institutional Review Board, Khon
Kaen University, Thailand (HE602158). Every participant provided prior
written consent.Detection of G6PD deficiency. EDTA blood samples were transported in a cold box to the laboratory within two hours of collection, stored at 4-8°C
and analyzed within one week of collection. Samples were initially
screened for G6PD activity by FST. The FST reagent was prepared
according to recommendations of the International Council (formerly
called Committee) for Standardization in Haematology (ICSH)[23] and stored at -20°C until used. FST was performed by adding 5 μL aliquot of whole blood to 100 μL of test reagent and 10 μL
aliquot of the mixture was immediately spotted onto Whatman No.1 filter
paper while the remaining solution was incubated at room temperature.
The second and third aliquot of the mixture was then placed onto the
same filter paper next to previous spot of the same sample after 5 and
15 minutes of incubation, respectively. Filter papers were dried and
examined visually under UV light (365 nm). A sample showing a moderate
to strong fluorescence at 5 minutes and strong fluorescence at 15
minutes was classified as G6PD normal, showing weak fluorescence at 5
minutes and moderate fluorescence at 15 minutes as G6PD intermediate
deficient and showing no or very faint fluorescence after 15 minutes as
G6PD deficient. FST assays were conducted alongside in-house G6PD
normal and intermediate G6PD deficient controls.G6PD
activity was quantified using a Trinity Biotech G6PD kit (Trinity
Biotech, Bray, Ireland). In brief, 10 μL aliquot of blood sample was
added to 90 μL of lysing reagent and mixed using a vortex mixer, and
hemolysate then was placed into a clinical chemistry analyzer (BS-400;
Mindray, Shenzhen, PR China) for measurement of G6PD activity and
hemoglobin (Hb) concentration. G6PD activity assay was based on NADP
reduction rate to NADPH catalyzed by G6PD as monitored at 37°C by an
increase in A340 nm measured at
regular intervals from 3 to 8 minutes post-addition of reagent mixture.
G6PD activity was calculated by multiplying the rate of increase in
absorbance (ΔA340 nm /min)
by 4,754 and expressed as U/g Hb. Each assay was accompanied by
parallel measurements of G6PD normal (11.3 ± 0.7 U/g Hb) and
intermediate (6.0 ± 0.2 U/g Hb) controls, with an inter-assay
coefficient of variation of 5.6 and 3.8%, respectively. Median G6PD
activity of all individuals carrying wild-type G6PD is defined as
normal G6PD activity, and G6PD status is classified as deficient,
intermediate deficient, and normal if the measured activity is <30,
30-70, and >70% normal activity, respectively.Genotyping of G6PD variants.Multiplex allele-specific (MAS)-PCR.
DNA was extracted from peripheral blood leukocytes using a DNAzol
reagent kit (Invitrogen, Carlsbad, CA, USA); MAS-PCR was performed
using primer sets specific for seven common G6PD variants in Southeast
Asia; namely, primer set 1 for G6PD Canton (c.1376G>T, p.R459L),
Union (c.1360C>T, p.R454C) and Viangchan (c.871G>A, p.V291M);
primer set 2 for G6PD Kaiping (c.1388G>A, p.R463H) and Mahidol
(c.487G>A, p.G163S); primer set 3 for G6PD Chinese-4 (c.392G>T,
p.G131V); and primer set 4 for G6PD Chinese-5 (c.1024C>T, p.L342F) (Table 1). Locations of primers on the G6PD gene are shown in Supplementary Figure S1.
Reaction mixture (50 μL) contained reaction buffer (10 mM Tris-HCl pH
8.3, 50 mM KCl, 3 mM MgCl2, and 0.001% gelatin), 200 mM dNTPs, 0.8 U
Taq DNA polymerase (New England BioLabs, Ipswich, MA, USA), primer set (Table 1), and 50 ng of extracted DNA. In each reaction mixture, primers GF1 and GR1 or GER4 (Table 1)
generating an internal control amplicon (339 or 825 bp respectively)
were also added. Thermocycling was carried out in a Biometra’s
T-personal Thermal Cycler (Biocompare, San Francisco, CA, USA) as
follows: 94°C for 3 minutes; followed by 30 cycles of 94°C for 60 s and
65°C (primer set 1), 70°C (primer sets 2 and 4) or 71°C (primer set 3)
for 90 s; and a final step of 16°C and maintained at this temperature
until analyzed by 2.0% agarose gel-electrophoresis, staining with
ethidium bromide and visualizing under UV light (Supplementary Figure S2).
MAS-PCR set 1 was carried out on all samples, and negative samples were
subsequently subjected to MAS-PCR assay using primer sets 2, 3 and 4.
All samples positive for G6PD variants were confirmed by RFLP-PCR as
previously described (data not shown).[24]
 |
Table 1. Primers used for detection of G-6-PD normal and mutant alleles.
|
DNA sequencing.
Samples classified by both FST and G6PD activity assay as G6PD
intermediate deficient but negative for any of the seven variants by
MAS-PCR as described above were then subjected to DNA sequencing.
Touchdown PCR reaction was performed to amplify the 13 exons of G6PD as
previously described.[25] Amplicons were purified
using a GenepHlow™ Gel/PCR Kit (Geneaid Biotech Ltd., Taipei, Taiwan),
directly sequenced (First BASE Lab., Selangor, Malaysia), and sequences
analyzed using a Seq-scanner program (Thermo Fisher Scientific,
Waltham, MA, USA).Statistical analysis.
Statistical analysis was carried out using a Minitab statistical
software version 14 (Minitab Inc., State College, PA, USA). Descriptive
statistics were applied to compare data and a GraphPad Prism6 (GraphPad
Software, San Diego, CA, USA) was employed to generate a G6PD variant
enzyme activity-distribution plot.
Results
The
overall prevalence of G6PD deficiency determined by FST was 18%, with
3.4% classified as deficient and 14.6% intermediate deficient, whereas
by G6PD activity assay, G6PD deficiency prevalence was 29.6%, with 4.5%
classified as deficient and 25.1% intermediate deficient (Table 2).
Genotypic tests identified 28.1% of women carrying G6PD variants, 2.2%
homozygotes or compound heterozygotes, and 25.9% heterozygotes (Table 2).
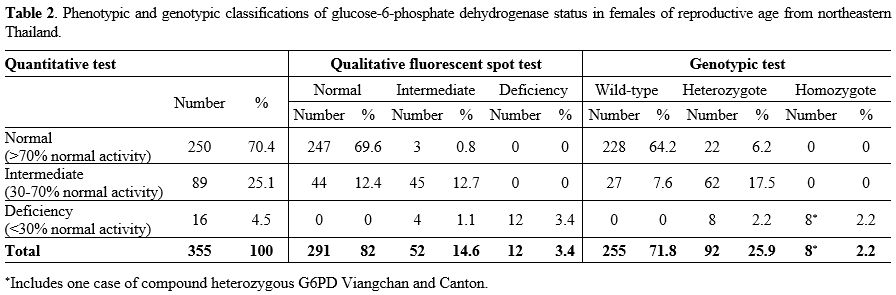 |
Table 2. Phenotypic and
genotypic classifications of glucose-6-phosphate dehydrogenase status
in females of reproductive age from northeastern Thailand.
|
All
homozygotes or compound heterozygotes were G6PD deficient (<30%
normal activity), but heterozygotes expressed a wide range of RBC G6PD
activity, 2.2% with enzyme activity in deficient range, 6.2% in normal
(>70% normal activity) range, and 17.5% in intermediate (30-70%
normal activity) range. The majority of individuals with wild-type G6PD alleles (64.2%) exhibited normal G6PD activity, although a minority (7.6%) expressed intermediate deficient activity.Intermediate
G6PD deficiency samples (n = 6), by both FST and enzyme activity assay,
and negative for PCR detection of the seven G6PD variants were
subjected to DNA sequencing, revealing two cases of heterozygous G6PD
Coimbra (c.592C>T, p.R198C), one case of heterozygous G6PD Gaohe
(c.95A>G, p.H32R), one case of homozygous silent mutation
(c.1311C>T) and two cases of a heterozygous novel mutation
(c.305T>C, p.F102S), named G6PD Khon Kaen (GenBank accession no.
MN316615). The moderately conserved residue F102 is located next to the
highly conserved residue F101 (Figure 1).
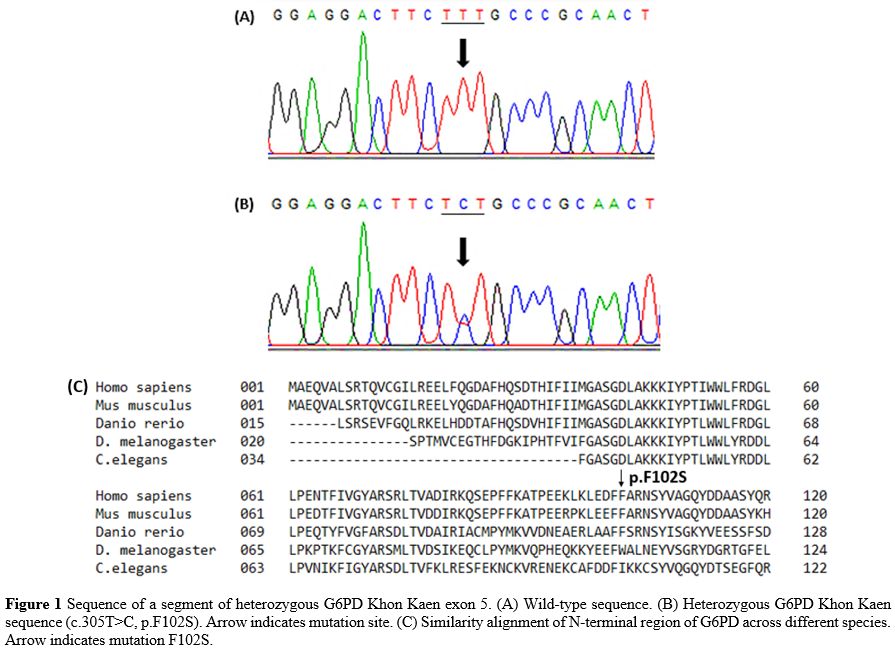 |
Figure 1. Sequence of a
segment of heterozygous G6PD Khon Kaen exon 5. (A) Wild-type sequence.
(B) Heterozygous G6PD Khon Kaen sequence (c.305T>C, p.F102S). Arrow
indicates mutation site. (C) Similarity alignment of N-terminal region
of G6PD across different species. Arrow indicates mutation F102S.
|
The
most common G6PD variant was Viangchan (54%), followed by Canton (11%)
and Union (11%). A case of Viangchan/Canton compound heterozygote was
also found. G6PD activity distribution demonstrated overlap of
activities among 92 heterozygotes and wild-type individuals (Figure 2).
It is worth noting that all non-common G6PD variants in Thailand (G6PD
Coimbra, Gaohe, and Khon Kaen) had low intermediate or deficient enzyme
activity (Figure 2).
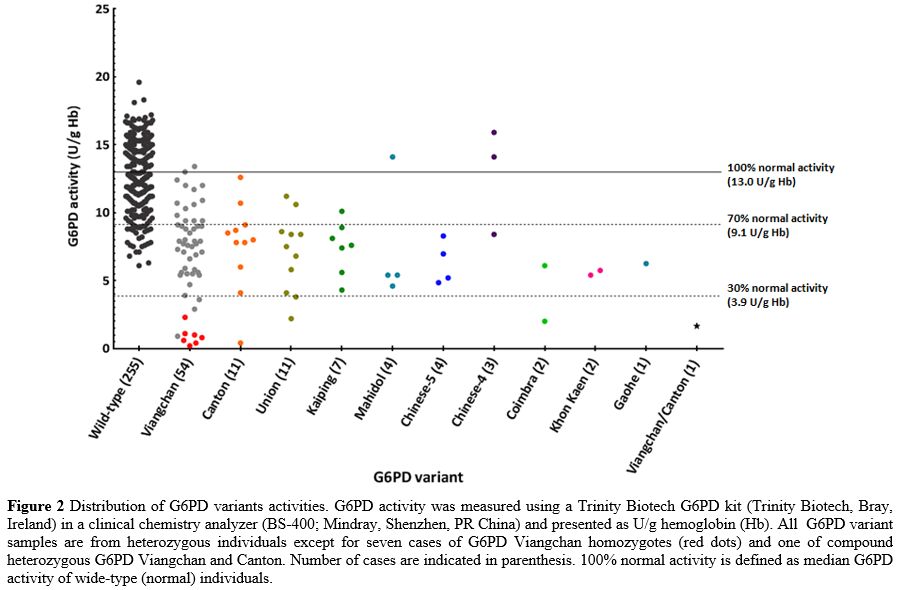 |
Figure 2. Distribution of
G6PD variants activities. G6PD activity was measured using a Trinity
Biotech G6PD kit (Trinity Biotech, Bray, Ireland) in a clinical
chemistry analyzer (BS-400; Mindray, Shenzhen, PR China) and presented
as U/g hemoglobin (Hb). All G6PD variant samples are from
heterozygous individuals except for seven cases of G6PD Viangchan
homozygotes (red dots) and one of compound heterozygous G6PD Viangchan
and Canton. Number of cases are indicated in parenthesis. 100% normal
activity is defined as median G6PD activity of wide-type (normal)
individuals.
|
Discussion
G6PD
deficiency is common in northeast Thailand. This is the first report of
an accurate determination of G6PD deficiency prevalence in northeastern
Thailand females by genotyping G6PD variants. The high prevalence of
G6PD deficiency in Thailand results from selective pressure of
malaria[26] as demonstrated in other parts of the world where malaria
is or has been endemic.[12,27]FST, the most commonly performed qualitative test, underestimates the prevalence of heterozygous G6PD deficiency;[28]
in the present study, about half of females with intermediate G6PD
activity were misclassified by FST having normal G6PD activity (Table 2). These falsely normal females are thus vulnerable to oxidative hemolysis. Furthermore, there were individuals carrying normal G6PD
alleles expressing intermediate G6PD activity based on enzyme activity
assay and heterozygotes presenting normal G6PD activity. Intermediate
G6PD activities found in normal G6PD individuals might result from a
high proportion of senescent RBCs in their circulation at the time of
blood collection, as G6PD activity decreases significantly according to
RBCs age.[29] In heterozygous G6PD-deficient females,
random X-chromosome inactivation results in various ratios of G6PD
normal to G6PD-deficient RBCs, thereby resulting in G6PD phenotypes
ranging from almost normal to severely deficient, while the majority
have G6PD activity in the intermediate range.[30]
Other factors impacting on G6PD activity of heterozygotes are enzyme
variant types and age of RBCs. Furthermore, rates of enzyme decay with
the aging of RBCs vary substantially among different G6PD variants.[31,32]
G6PD variants identified in this study belong to class II (Canton,
Coimbra, Kaiping, Union, and Viangchan) and Class III (Chinese-4,
Chinese-5, Gaohe and Mahidol).[2,33]
No clear differences in G6PD activity between the two enzyme classes
could be discerned. Moreover, median G6PD activity (defined as normal
activity) in this study is calculated from enzyme activity of females
with wild-type G6PD alleles and not from males, and the value was
slightly higher than those of previous reports.[6] In
northeastern Thailand, about 50% of women of reproductive age carry
various thalassemia genes resulting in significantly higher
reticulocyte count, especially immature reticulocyte fraction (IRF),[34]
and higher G6PD activity. In addition, the non-common G6PD variants in
this region (G6PD Coimbra, Gaohe and Khon Kaen) had low intermediate or
deficient enzyme activity; therefore, they will be assuredly diagnosed
by G6PD activity assay.In this region of Thailand, G6PD Viangchan accounts for >50% of the seven common variants.[24] G6PD Viangchan is also the most common variant in Cambodia and PDR Laos,[35,36] reflecting the common ancestral origin of populations in northeastern Thailand, Cambodia and PDR Laos.[24,37]
The other less common variants found in this study were Canton,
Chinese-4, Chinese-5, Gaohe, Kaiping and Union (common variants in
Chinese population), and Mahidol (the most common variant in Myanmar
population),[33] indicating the presence of people originating from these two countries in northeastern Thailand. The
novel G6PD Khon Kaen (c.305T>C, p.F102S) was discovered among
samples with intermediate G6PD deficiency. The F102 residue is located
within the coenzyme-binding domain (residues 31-200).[38]
Replacement of phenylalanine with a smaller and more hydrophilic serine
residue would be sufficient to cause impaired folding of the domain,
leading to decreased stability and low enzymatic activity.[39]
Conclusions
The
results highlight the high prevalence in northeastern Thailand of
females heterozygous for G6PD variants, the majority with G6PD activity
in the intermediate deficiency range; however, some of these
samples show normal and deficient enzyme activity ranges, indicating
that the accurate identification of female G6PD deficiency carriers
requires both measurements of G6PD activity and genotyping of G6PD
variants. Knowing the genotype-phenotype correlation of G6PD deficiency
in this female population will help improve the management of acute
hemolysis related to drugs, foods, and infections.
Acknowledgements
The
research was supported by Khon Kaen University, Thailand (grant number
600068) and the Faculty of Associated Medical Sciences, Khon Kaen
University, Thailand. We thank Professor Dr. Prapon Wilairat for
valuable suggestions and editing the first draft of the manuscript.
References
- Luzzatto L, Ally M, Notaro R. Glucose-6-phosphate dehydrogenase deficiency. Blood. 2020;136(11):1225-1240. https://doi.org/10.1182/blood.2019000944 PMid:32764983 PMCid:PMC7371606
- WHO Working Group. Glucose-6-phosphate dehydrogenase deficiency. Bull World Health Organ. 1989;67(6):601-611. https://apps.who.int/iris/handle/10665/264721 PMid:2633878 PMCid:PMC2491315
- World
Health Organization. Guide to G6PD deficiency rapid diagnostic testing
to support P. vivax radical cure. Geneva, World Heal Organ. 2018. https://www.who.int/publications/i/item/9789241514286
- Commons
RJ, McCarthy JS, Price RN. Tafenoquine for the radical cure and
prevention of malaria: the importance of testing for G6PD deficiency.
Med J Aust. 2020;212(4):152-153.el. https://doi.org/10.5694/mja2.50474 PMid: 32036613 PMCid: PMC7064913
- Minucci
A, Giardina B, Zuppi C, Capoluongo E. Glucose-6-phosphate dehydrogenase
laboratory assay: How, when, and why? IUBMB Life. 2009;61(1):27-34. https://doi.org/10.1002/iub.137 PMid:18942156
- Pfeffer
DA, Ley B, Howes RE, Adu P, Alam MS, Bansil P, Boum 2nd Y, Brito M,
Charoenkwan P, Clements A, Cui L, Deng Z, Egesie OJ , Espino FE,
Fricken ME, Hamid MMA, He Y, Henriques G, Khan WA, Khim N, Kim S,
Lacerda M, Lon C, Mekuria AH, Menard D, Monteiro W, Nosten F, Oo NN,
Pal S, Palasuwan D, Parikh S, Pasaribu AP, Poespoprodjo JR, Price DJ,
Roca-Feltrer A, Roh ME, Saunders DL, Spring MD, Sutanto I,
Thriemer K, Weppelmann TA, Seidlein L, Satyagraha AW, Bancone G,
Domingo GJ, Price RN. Quantification of glucose-6-phosphate
dehydrogenase activity by spectrophotometry: a systematic review and
meta-analysis. PLoS Med. 2020;17(5). https://doi.org/10.1371/journal.pmed.1003084 PMid:32407380 PMCid:PMC7224463
- Kaplan
M, Beutler E, Vreman HJ, Hammerman C, Levy-Lahad E, Renbaum P,
Stevenson DK. Neonatal hyperbilirubinemia in glucose-6-phosphate
dehydrogenase-deficient heterozygotes. Pediatrics. 1999;104:68-74. https://doi.org/10.1542/peds.104.1.68 PMid:10390262
- Zangen
S, Kidron D, Gelbart T, Roy-Chowdhury N, Wang X, Kaplan M. Fatal
kernicterus in a girl deficient in glucose-6-phosphate dehydrogenase: a
paradigm of synergistic heterozygosity. J Pediatr. 2009;154(4):616-619.
https://doi.org/10.1016/j.jpeds.2008.10.049 PMid:19324225
- Chu
CS, Bancone G, Nosten F, White NJ, Luzzatto L. Primaquine-induced
haemolysis in females heterozygous for G6PD deficiency. Malar J.
2018;17(1):101. https://doi.org/10.1186/s12936-018-2248-y PMid:29499733 PMCid:PMC5833093
- Chu CS, Freedman DO. Tafenoquine and G6PD: a primer for clinicians. J Travel Med. 2019;26(4):1-11. https://doi.org/10.1093/jtm/taz023 PMid:30941413 PMCid:PMC6542331
- Luzzatto
L, Seneca E. G6PD deficiency: a classic example of pharmacogenetics
with on-going clinical implications. Br J Haematol.
2014;164(4):469-480. https://doi.org/10.1111/bjh.12665 PMid: 24372186 PMCid: PMC4153881
- Howes
RE, Battle KE, Satyagraha AW, Baird JK, Hay SI. G6PD deficiency: global
distribution, genetic variants and primaquine therapy. Adv Parasitol.
2013;81:133-201. https://doi.org/10.1016/B978-0-12-407826-0.00004-7 PMid: 23384623
- Petney
T, Sithithaworn P, Satrawaha R, Warr CG, Andrews R, Wang YC, Feng CC.
Potential malaria reemergence, northeastern Thailand. Emerg Infect Dis.
2009;15(8):1330-1331. https://doi.org/10.3201/eid1508.090240 PMid:9751612 PMCid:PMC2815977
- Imwong
M, Dhorda M, Myo Tun K, Thu AM, Phyo AP, Proux S, Suwannasin K, Kunasol
C, Srisutham S, Duanguppama J, Vongpromek R, Promnarate C, Saejeng A,
Khantikul N, Sugaram R, Thanapongpichat S, Sawangjaroen N, Sutawong K,
Han KT, Htut Y, Linn K, Win AA, Hlaing TM, van der Pluijm RW,
Mayxay M, Pongvongsa T, Phommasone K, Tripura R, Peto TJ, von Seidlein
L, Nguon C, Lek D, Chan XHS, Rekol H, Leang R, Huch C, Kwiatkowski DP,
Miotto O, Ashley EA, Kyaw MP, Pukrittayakamee S, Day NPJ, Dondorp AM,
Smithuis FM, Nosten FH, White NJ. Molecular epidemiology of resistance
to antimalarial drugs in the Greater Mekong subregion: an observational
study. Lancet Infect Dis. 2020;20(12):1470-1480. https://doi.org/10.1016/S1473-3099(20)30228-0 PMid:32679084
- Geng
J, Malla P, Zhang J, Xu S, Li C, Zhao Y, Wang Q, Kyaw MP, Cao Y, Yang
Z, Cui L. Increasing trends of malaria in a border area of the Greater
Mekong Subregion. Malar J. 2019;18:309. https://doi.org/10.1186/s12936-019-2924-6 PMid:31514740 PMCid:PMC6739967
- Wasi
P, Na-Nakorn S, Suingdumrong A. Studies of the distribution of
haemoglobin E, thalassaemias and glucose-6-phosphate dehydrogenase
deficiency in North-eastern Thailand. Nature. 1967;214(5087):501-502. https://doi.org/10.1038/214501a0 PMid:6032880
- Kruatrachue
M, Charoenlarp P, Chongsuphajaisiddhi T, Harinasuta C. Erythrocyte
glucose-6-phosphate dehydrogenase and malaria in Thailand. Lancet.
1962;2(7267):1183-1186. https://doi.org/10.1016/s0140-6736(62)90956-x PMid:13927068
- Flatz G, Sringam S. Malaria and glucose-6-phosphate dehydrogenase deficiency in Thailand. Lancet. 1963;282(7320):1248-1250. https://doi.org/10.1016/s0140-6736(63)90895-x PMid:14066844
- Flatz
G, Sringam S. Glucose-6-phosphate dehydrogenase deficiency in different
ethnic groups in Thailand. Ann Hum Genet. 1964;27:315-318. https://doi.org/10.1111/j.1469-1809.1963.tb01528.x PMid:14175196
- Tuchinda
S, Rucknagel DL, Na-Nakorn S, Wasi P. The Thai variant and the
distribution of alleles of 6-phosphogluconate dehydrogenase and the
distribution of glucose 6-phosphate dehydrogenase deficiency in
Thailand. Biochem Genet. 1968;2(3):253-264. https://doi.org/10.1007/BF01474765 PMid:5715190
- Kittiwatanasarn
P, Louicharoen C, Sukkapan P, Nuchprayoon I. Glucose-6-phosphate
dehydrogenase deficiency in Northeastern Thailand: prevalence and
relationship to neonatal jaundice. Chula Med J. 2003;47(8):471-479.
- Jamnok
J, Sanchaisuriya K, Sanchaisuriya P, Fucharoen G, Fucharoen S, Ahmed F.
Factors associated with anaemia and iron deficiency among women of
reproductive age in Northeast Thailand: a cross-sectional study. BMC
Public Health. 2020;20:102. https://doi.org/10.1186/s12889-020-8248-1 PMid:31992253 PMCid:PMC6986100
- Beutler
E, Blume KG, Kaplan JC, Löhr GW, Ramot B, Valentine WN. International
Committee for Standardization in Haematology: recommended screening
test for glucose-6-phosphate dehydrogenase (G-6-PD) deficiency. Br J
Haematol. 1979;43(3):465-467. https://doi.org/10.1111/j.1365-2141.1979.tb03774.x PMid:497122
- Kitcharoen
S, Dechyotin S, Khemtonglang N, Kleesuk C. Relationship among
glucose-6-phosphate dehydrogenase (G-6-PD) activity, G-6-PD variants
and reticulocytosis in neonates of northeast Thailand. Clin Chim Acta.
2015;442:125-129. https://doi.org/10.1016/j.cca.2015.01.017 PMid:25632835
- Dallol
A, Banni H, Gari MA, Al-Qahtani MH, Abuzenadeh AM, Al-Sayes F,
Chaudhary AG, Bidwell J, Kafienah W. Five novel glucose-6-phosphate
dehydrogenase deficiency haplotypes correlating with disease severity.
J Transl Med. 2012;10:199. https://doi.org/10.1186/1479-5876-10-199 PMid:23006493 PMCid:PMC3492101
- Poolsuwan S. Testing the “malaria hypothesis”for the case of Thailand: a genetic appraisal. Hum Biol. 2003;75(4):585-605. https://doi.org/10.1353/hub.2003.0061 PMid:14655879
- Siniscalco
M, Bernini L, Filippi G, Latte B, Meera Khan P, Piomelli S, Rattazzi M.
Population genetics of haemoglobin variants, thalassaemia and
glucose-6-phosphate dehydrogenase deficiency, with particular reference
to the malaria hypothesis. Bull World Health Organ.1966;34(3):379-93.
PMid: 5296398 PMCid:PMC 2475975
- Chu CS,
Bancone G, Kelley M, Advani N, Domingo GJ, Cutiongo-de la Paz EM, van
der Merwe N, Cohen J, Gerth-Guyette E. Optimizing G6PD testing for
Plasmodium vivax case management and beyond: why sex, counseling, and
community engagement matter. Wellcome Open Res. 2020;5:21. https://doi.org/10.12688/wellcomeopenres.15700.2 PMid:32766454 PMCid:PMC7388194
- Marks
PA, Johnson AB, Hirschberg E. Effect of age on the enzyme activity in erythrocytes. Proc Natl Acad Sci. 1958;44(6):529-536. https://doi.org/10.1073/pnas.44.6.529 PMid:16590234
- Beutler
E, Yeh M, Fairbanks VF. The normal human female as a mosaic of
X-chromosome activity: studies using the gene for G-6-PD-deficiency as
a marker. Proc Natl Acad Sci U S A. 1962;48(1):9-16. https://doi.org/10.1073/pnas.48.1.9 PMid: 13868717
- Morelli
A, Benatti U, Gaetani GF, De Flora A. Biochemical mechanisms of
glucose-6-phosphate dehydrogenase deficiency. Proc Natl Acad Sci U S A.
1978;75(4):1979-1983. https://doi.org/10.1073/pnas.75.4.1979 PMid: 273924 PMCid: PMC392466
- Piomelli
S, Corash LM, Davenport DD, Miraglia J, Amorosi EL. In vivo lability of
glucose-6-phosphate dehydrogenase in GdA- and GdMediterranean
deficiency. J Clin Invest. 1968;47(4):940-948. https://doi.org/10.1172/JCI105786 PMid: 5641629 PMCid: PMC297242
- Minucci
A, Moradkhani K, Hwang MJ, Zuppi C, Giardina B, Capoluongo E.
Glucose-6-phosphate dehydrogenase (G6PD) mutations database: review of
the “old” and update of the new mutations. Blood Cells Mol Dis.
2012;48(3):154-165. https://doi.org/10.1016/j.bcmd.2012.01.001 PMid: 22293322
- Jamnok
J, Sanchaisuriya K, Chaitriphop C, Sanchaisuriya P, Fucharoen G,
Fucharoen S. A new indicator derived from reticulocyte hemoglobin
content for screening iron deficiency in an area prevalent for
thalassemia. Lab Med. 2020;51(5):498-506. https://doi.org/10.1093/labmed/lmz099 PMid: 32052840
- Matsuoka
H, Nguon C, Kanbe T, Jalloh A, Sato H, Yoshida S, Hirai M, Arai M,
Socheat D, Kawamoto F. Glucose-6-phosphate dehydrogenase (G6PD)
mutations in Cambodia: G6PD Viangchan (871G>A) is the most common
variant in the Cambodian population. J Hum Genet. 2005;50(9):468-472. https://doi.org/10.1007/s10038-005-0279-z PMid:16136268
- Bancone
G, Menard D, Khim N, Kim S, Canier L, Nguong C, Phommasone K, Mayxay M,
Dittrich S, Vongsouvath M, Fievet N, Le Hesran JY, Briand V, Keomany S,
Newton PN, Gorsawun G, Tardy K, Chu CS, Rattanapalroj O, Dong LT, Quang
HH, Tam-Uyen N, Thuy-Nhien N, Hien TT, Kalnoky M, Nosten F. Molecular
characterization and mapping of glucose-6-phosphate dehydrogenase
(G6PD) mutations in the Greater Mekong Subregion. Malar J. 2019;18:20. https://doi.org/10.1186/s12936-019-2652-y PMid:30674319 PMCid:PMC6343352
- Laosombat
V, Sattayasevana B, Janejindamai W, Viprakasit V, Shirakawa T,
Nishiyama K, Matsuo M. Molecular heterogeneity of glucose-6-phosphate
dehydrogenase (G6PD) variants in the south of Thailand and
identification of a novel variant (G6PD Songklanagarind). Blood Cells
Mol Dis. 2005;34(2):191-196. https://doi.org/10.1016/j.bcmd.2004.11.001 PMid:15727905
- Au
SWN, Gover S, Lam VMS, Adams MJ. Human glucose-6-phosphate
dehydrogenase: The crystal structure reveals a structural NADP+
molecule and provides insights into enzyme deficiency. Structure.
2000;8:293-303. https://doi.org/10.1016/s0969-2126(00)00104-0 PMid:10745013
- Cunningham
AD, Colavin A, Huang KC, Mochly-Rosen D. Coupling between protein
stability and catalytic activity determines pathogenicity of G6PD
variants. Cell Rep. 2017;18(11):2592-2599. https://doi:10.1016/j.celrep.2017.02.048 PMid:28297664
Supplementary files
 |
Supplementary Figure S1. Location on glucose-6-phosphate dehydrogenase gene (G6PD)
of allele-specific primers for multiplex detection of G6PD Canton,
Union and Viangchan (Set 1), G6PD Kaiping and Mahidol (Set 2), G6PD
Chinese-4 (Set 3) and G6PD Chinese-5 (Set 4). Location of internal
control primers are also indicated. Primer sequences are listed in
Table 1.
|
 |
Supplementary Figure S2
Agarose gel-electrophoresis of multiplex allele-specific PCR amplicons
of seven common heterozygous glucose-6-phosphate (G6PD) variants
present in Southeast Asia. (A) Lane M, 100 bp DNA size markers; lane 1,
uniplex internal control primers (IC); lane 2, G6PD Viangchan (VC)
allele plus IC; lane 3, G6PD Canton (CT) allele plus IC; lane 4, G6PD
Union (UN) allele plus IC. (B) Lane M, 100 bp DNA size markers; lane 1,
uniplex internal control primers (IC); lane 2, G6PD Kaiping (KP) allele
plus IC; lane 3, G6PD Mahidol (MH) allele plus IC. (C) Lane M, 100 bp
DNA size markers; lane 1, uniplex internal control primers (IC); lane
2, G6PD Chinese-4 (C-4) plus IC. (D) Lane M, 100 bp DNA size markers;
lane 1, uniplex internal control primers (IC); lane 2, G6PD Chinese-5
(C-5) allele plus IC. (E) Lane M, 50 bp DNA size markers; lane 2, 4 and
6, uniplex internal control primers (IC); lane 1, normal allele (VC_N)
present in heterozygous VC sample plus IC; lane 3, normal allele (CT_N)
present in heterozygous CT sample plus IC; lane 5, normal allele (UN_N)
present in heterozygous UN sample plus IC. (F) Lane M, 50 bp DNA size
markers; lane 2, 4, 6, and 8, uniplex internal control primers (IC);
lane 1, normal allele (KP_N) present in heterozygous KP sample plus IC;
lane 3, normal allele (MH_N) present in heterozygous MH sample plus IC;
lane 5, normal allele (C-4_N) present in heterozygous C-4 sample plus
IC; lane 7, normal allele (C-5_N) present in heterozygous C-5 sample
plus IC.
|
[TOP]



