Feiying
Meng1,2.
1
State Key Laboratory of Experimental Hematology, Tianjin, 300020, P. R.
China.
2
Henan Provincial People’s Hospital, Henan Eye Hospital, Henan Eye
Institute, People’s Hospital of Zhengzhou University, Zhengzhou, Henan
450003, P. R. China.
Correspondence to: Feiying
Meng, State Key Laboratory of Experimental Hematology, Tianjin, 300020,
P.R.China; Henan Provincial People’s Hospital, Henan Eye Hospital,
Henan Eye Institute, People’s Hospital of Zhengzhou University,
Zhengzhou, Henan 450003, P.R.China. Tel: 86-19139761276. E-mail:
fymeng32@163.com
Published: July 1, 2021
Received: January 15, 2021
Accepted: June 7, 2021
Mediterr J Hematol Infect Dis 2021, 13(1): e2021041 DOI
10.4084/MJHID.2021.041
This is an Open Access article distributed
under the terms of the Creative Commons Attribution License
(https://creativecommons.org/licenses/by-nc/4.0),
which permits unrestricted use, distribution, and reproduction in any
medium, provided the original work is properly cited.
|
|
Abstract
Objective: This
study aims at uncovering the effects of microRNAs (miRNAs) on the F8 gene and FVIII
protein in hemophilia A (HA).
Methods: F8-targeting
miRNAs were predicted by TargetScan, miRDB, and starBase. MiRNAs,
predicted by at least two of the three databases, were selected for
further study, and their expressions in the blood of HA patients
without F8
mutations and
healthy controls were detected. A dual-luciferase reporter assay was
performed to verify the binding between hsa-miR-5581-3p/hsa-miR-542-3p
and F8. In
addition, the regulation of F8
by hsa-miR-5581-3p/hsa-miR-542-3p was investigated in human umbilical
vein endothelial cells (HUVECs) and lymphoblastoid cell line (LCL) that
displayed endogenous expression of FVIII. qRT-PCR was used to detect
the expressions of miRNAs and F8
gene, and Western blotting was conducted to measure the expression of
FVIII protein.
Results: A total of 42 F8-targeting
miRNAs were predicted by at least two of the three databases. Among
these miRNAs, hsa-miR-5581-3p and hsa-miR-542-3p were highly expressed
in the blood of HA patients and have not been reported in previous
studies of HA. Both hsa-miR-5581-3p and hsa-miR-542-3p could bind the
3’UTR of F8
mRNA. Upregulation of hsa-miR-5581-3p or hsa-miR-542-3p suppressed the
expressions of F8
mRNA and FVIII protein in HUVECs and LCL cells.
Conclusion: Hsa-miR-5581-3p and hsa-miR-542-3p target the F8 gene and
suppress the expression of FVIII protein, which may contribute to the
development of HA without F8
mutations.
|
Introduction
Hemophilia
is a hereditary bleeding disorder resulting from deficiency or
dysfunction of coagulation protein factor VIII (FVIII) and IX (FIX),
leading to hemophilia A (HA) and hemophilia B (HB), respectively. The
prevalence of HA is about fourfold higher than that of HB, and the
prevalence of severe hemophilia is about 37% of all cases.[1]
Severe hemophilia is manifested by spontaneous muscle or intraarticular
hemorrhage, and recurrent joint hemorrhages can result in progressive
tissue damage and joint.[2]
Prophylactic factor
replacement is currently the standard treatment option for hemophilia,
which is dependent on frequent intravenous injections.[3]
However, among a list of novel treatment strategies for hemophilia,
gene therapy shows a great appeal due to its potential for endogenous
production of FVIII or FIX by transferring a functional gene to replace
the deficient gene.[4]
In HA, the deficiency of FVIII is mainly caused by mutations in the F8 gene. Intron 22
and intron 1 inversions are the most frequent F8 variants in HA
patients, followed by recurrent mutation at nucleotide 6046
(c.6046C>T) in the F8
gene.[5]
Missense mutations in this region (c.6046C>T/p.R2016W) damage
secretion and activity of FVIII and also impede the correct pre-mRNA
splicing, which further impairs FVIII activity.[6]
Despite clinically reportable DNA variants in 98.1% of HA patients,[7] a small portion of HA patients is not
affected by F8
variants, which encourages us to find the mechanism for
mutation-independent FVIII deficiency.
MicroRNAs
(miRNAs) are the most-investigated class of small non-coding RNAs
(ncRNAs) for their ubiquitous regulation in the human genome. MiRNAs
mainly regulate gene expression in cell cytoplasm in a
sequence-specific manner by recruiting an RNA-induced silencing complex
to their target RNAs.[8] This
miRNA-target interaction
results in mRNA degradation or translational repression according to
the degree of complementarity.[9]
Numerous functional studies have proved a causal link between miRNA
dysregulation and human diseases, particularly cancers.
Rosset et al. firstly found a single nucleotide polymorphism in the
3’UTR of F8
(c.8728A>G, rs1050705) contributed to the complementarity
between F8
3’UTR and the seed sequences of miR-26a-5p and miR-26b-5p, which
further modified the FVIII residual clotting activity in HA patients.[10] A previous study reported that
miR-1246 was upregulated in the whole blood of HA patients, and this
ncRNA could suppress F8
expression by binding to the non-coding region of F8 mRNA.[11] Jankowska et al. identified
miR-30c-5p and miR-374b-5p as direct mediators of F8 gene and FVIII
in HA patients with normal F8
genotypes.[12] They also found
that murine miRNAs, including miR-208a, miR-351 and miR-125a, could
target the 3’UTR of F8
mRNA in mouse models and mammalian cells.[13]
The above evidence strongly supports the involvement of miRNAs in the
pathogenesis of HA.
The author applied three online databases to predict the F8-binding miRNAs
and detected their expressions in the blood of HA patients without F8 gene mutations.
Among the upregulated F8-binding
miRNAs, hsa-miR-5581-3p and hsa-miR-542-3p were selected for further
investigations since they were not reported in previous studies of HA.
The function of these two miRNAs was experimentally verified.
Materials and Methods
Prediction
of F8-targeting miRNAs.
F8-targeting
miRNAs were predicted by TargetScan (http://www.targetscan.org/vert_72/),
miRDB (http://www.mirdb.org/)
and starBase (http://starbase.sysu.edu.cn/index.php).
MiRNAs predicted by at least two of the three databases were selected
for further investigations.
Blood samples.
Blood samples were taken from two HA patients (one mild and one
moderate) without mutations in the F8
gene and three healthy controls. The participants were enrolled at the
Hematology Department of Henan Provincial People’s Hospital. Tubes
containing sodium citrate anticoagulant were used for the blood
collection. The blood samples were mixed with RNAlater Solution
(Qiagen, Valencia, CA, USA) and stored at -20°C until RNA isolation.
Mutations in intron 1/22 and all exons of the F8
gene were analyzed by next-generation sequencing and determined by
comparison with the reference (NG_011403.1). Both the two HA patients
had low FVIII activity (4.7 IU/dL and 19 IU/dL), but the RNA sequencing
showed no mutations in the exon and intron of the F8
gene in these two patients. Moreover, the level of vWF:Ag and the
activity of other coagulation factors, including FII, FV, FVII, FIX,
and FX, remained unchanged in these two HA patients. Mutations of the
vWF, LMAN1, and MCFD2 genes were excluded. The collection and use of
the blood samples obtained informed consent from all the participants
and approval of the ethics committee of Henan Provincial People’s
Hospital.
qRT-PCR.
A GenElute™ Plasma/Serum RNA Purification Mini Kit (Sigma-Aldrich, St.
Louis, MO, USA) and TRIzol reagent (Invitrogen, Carlsbad, CA, USA) were
used for total RNA extraction from blood samples and cells,
respectively. Extracted RNA was reverse-transcribed using a reverse
transcription kit (TaKaRa, Tokyo, Japan) based on the user’s manual.
Gene expression was detected by LightCycler 480 (Roche Diagnostics,
Indianapolis, IN, USA) under the conditions provided by the SYBR Green
Mix (Roche Diagnostics). The thermal cycling consisted of 10 s at 95°C,
45 cycles of 5 s at 95°C, 10 s at 60°C and 10 s at 72°C, and a final
extension for 5 min at 72°C. PCR test of each sample was duplicated
three times. U6 and GAPDH were used as the internal references of miRNA
and mRNA, respectively. Data were analyzed using the 2-ΔΔCt
method. ΔΔCt = (Ct target gene-Ct
reference gene)
experimental group-(Ct
target gene-Ct
reference gene)
control group.
Primers used for PCR amplification and their sequences are presented in
Table 1.
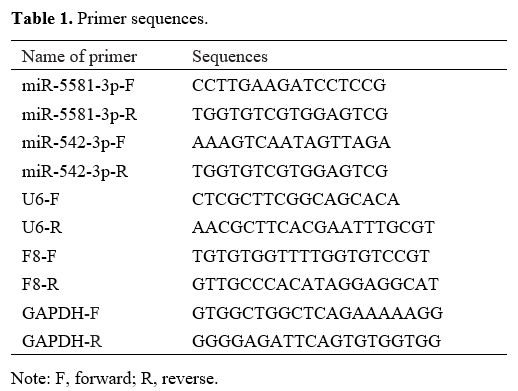 |
Table
1. Primer sequences.
|
Dual-luciferase
reporter assay.
Wild and mutant sequences of the binding sites of F8 (wt-F8 and mut-F8)
were designed and synthesized according to the online prediction. The
sequence contained the 3'UTR fragment complementary to the target
miRNA. Wt-F8 or mut-F8
was inserted into pGL3-Promoter vector and cotransfected with 50 nM
hsa-miR-5581-3p mimic, hsa-miR-542-3p mimic or mimic NC together with
Renilla luciferase reporter vector pRL-TK into HEK293T cells. The cells
were added with 100 µl of lysis buffer and placed on a shaker for 20
min at room temperature. Suspension of the cell lysates (50 µl) was
added with luciferase reaction solution (50 µl, Promega, Madison, WI,
USA) before Firefly luciferase activity was measured. The suspension
was then mixed with 50 µl of Stop&Glo reagent (Promega) to
measure
Renilla luciferase activity (internal reference). The ratio of firefly
luciferase activity to Renilla luciferase activity was the relative
luciferase activity.
Cell culture.
Human umbilical vein endothelial cells (HUVECs) and lymphoblastoid cell
line (LCL) that displayed endogenous expression of FVIII, and human
embryonic kidney cells (HEK293T) were acquired from the American Type
Culture Collection (ATCC, Manassas, Virginia, USA). HUVECs were
cultured in an endothelial basal medium (EBM-2, PromoCell, Heidelberg,
Germany) containing the Supplement Mix (PromoCell). HEK293T cells were
cultured in DMEM (Gibco, Grand Island, NY, USA) and LCL cells were in
RPMI medium (Gibco). The media for HEK293T and LCL cells were
supplemented with 10% fetal bovine serum, 100 U/ml penicillin and 100
mg/ml streptomycin. The cells were cultured at 37°C with 5% CO2.
Cell transfection.
HUVECs and LCL cells were transfected with hsa-miR-5581-3p mimic,
hsa-miR-542-3p mimic, hsa-miR-5581-3p inhibitor, hsa-miR-542-3p
inhibitor or relevant negative controls (NC) (50 nM, GenePharma,
Shanghai, China) using Lipofectamine 2000 reagent (Invitrogen,
Carlsbad, CA, USA). The cells were transfected in a 6-well plate
containing serum-free medium, and each well was seeded with 2 × 105 cells. Cells
with over 70% transfection efficiency were selected 24 h after the
transfection for the following experiments.
Western blotting.
Cells were treated with RIPA lysis buffer (Beyotime, Shanghai, China)
for protein extraction. Protein concentration was measured using a BCA
kit (Beyotime). Protein samples were added into reducing SDS-PAGE
loading buffer and boiled for 3 min. The proteins were electrophoresed
at 80 V for 30 min. After bromophenol blue entered the separation gel,
the electrophoresis was shifted to 120 V and maintained for 1 ~ 2 h.
The proteins were transferred to a membrane in an ice bath. The protein
transfer was carried out for 60 min with a current of 300 mA. The
membrane was immersed in blocking buffer at room temperature for 60 min
or at 4°C overnight. Primary antibodies against GAPDH (ab8245, 1:5000,
Abcam, Cambridge, MA, USA) and FVIII (ab171825, 1:1000, Abcam) were
incubated with the membrane on a shaker for 1 h at room temperature,
followed by incubation of secondary antibody for 1 h at room
temperature. The membrane was added with a color-developing solution
and subjected to protein expression analysis. Recombinant FVIII protein
antibody (ab158403, Abcam) was used as a positive control to detect
FVIII protein. The bands were quantified by Image J software (version
1.46, National Institutes of Health).
Statistical analysis. GraphPad
Prism 7 was used for statistical analysis. All data were expressed as
mean ± standard deviation. T-test
and One-way analysis of variance were used for two-group and multigroup
comparisons, respectively. Tukey's test was conducted for post hoc
multiple comparisons. Each experiment was repeated three times. P < 0.05 was
considered statistically significant.
Results
F8-targeting
miRNAs.
TargetScan, miRDB, and starBase predicted miRNAs that targeted the F8 gene. Among
these F8-targeting
miRNAs, hsa-miR-5581-3p was predicted by all the three databases, and
41 miRNAs were predicted by two of the three databases (Figure 1A). qRT-PCR
was used to detect the expressions of these 42 miRNAs (Supplementary Table 1)
in the blood of two HA patients without F8
mutations and three healthy controls. The experiment results showed
higher expressions of hsa-miR-5581-3p, hsa-miR-30c-3p, hsa-miR-374b-3p,
hsa-miR-542-3p and hsa-miR-6803-3p in HA patients than in healthy
controls (Figure 1B,
P <
0.05). Since hsa-miR-5581-3p and hsa-miR-542-3p have not been reported
in previous studies of hemophilia without F8 mutations, these
two miRNAs were selected for further investigations in our study.
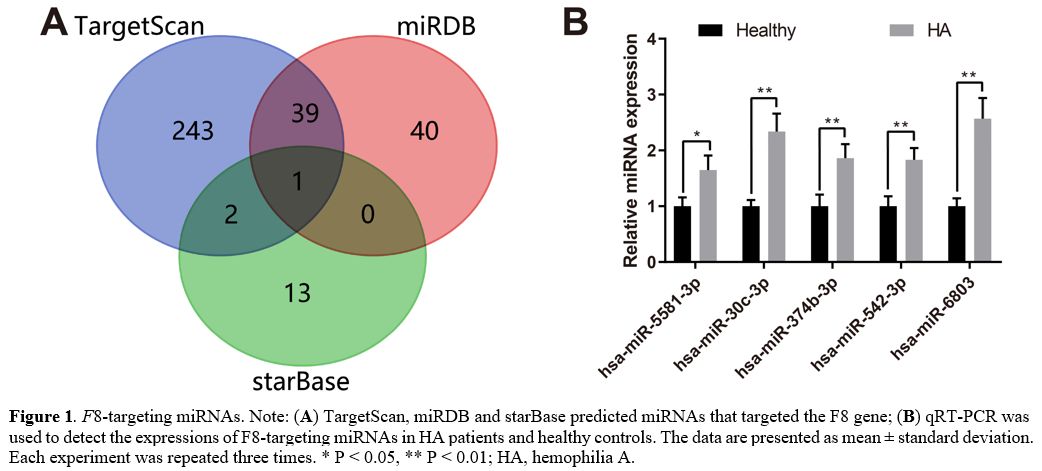 |
Figure
1. F8-targeting
miRNAs. Note: (A)
TargetScan, miRDB and starBase predicted miRNAs that targeted the F8 gene; (B) qRT-PCR was used
to detect the expressions of F8-targeting
miRNAs in HA patients and healthy controls. The data are presented as
mean ± standard deviation. Each experiment was repeated three times. * P < 0.05, **
P
< 0.01; HA, hemophilia A.
|
F8
is a target gene of hsa-miR-5581-3p and hsa-miR-542-3p. The
binding sites between F8 and
hsa-miR-5581-3p/hsa-miR-542-3p and the designed mutations are presented
in Figure 2A.
A dual-luciferase reporter assay was performed to verify the binding
between F8
and hsa-miR-5581-3p/hsa-miR-542-3p. HEK293T cells were transfected with
hsa-miR-5581-3p mimic, hsa-miR-542-3p mimic or mimic NC together with
luciferase reporter plasmids inserted with wt-F8 or mut-F8. Transfection
of wt-F8 +
hsa-miR-5581-3p mimic or hsa-miR-542-3p mimic reduced the luciferase
activity in the HEK293T cells (Figure
2B-C, P
< 0.05), while the luciferase activity in HEK293T cells
transfected with mut-F8 +
hsa-miR-5581-3p/hsa-miR-542-3p mimic remained unchanged. These results
validated the prediction that F8 was targeted
by hsa-miR-5581-3p and hsa-miR-542-3p.
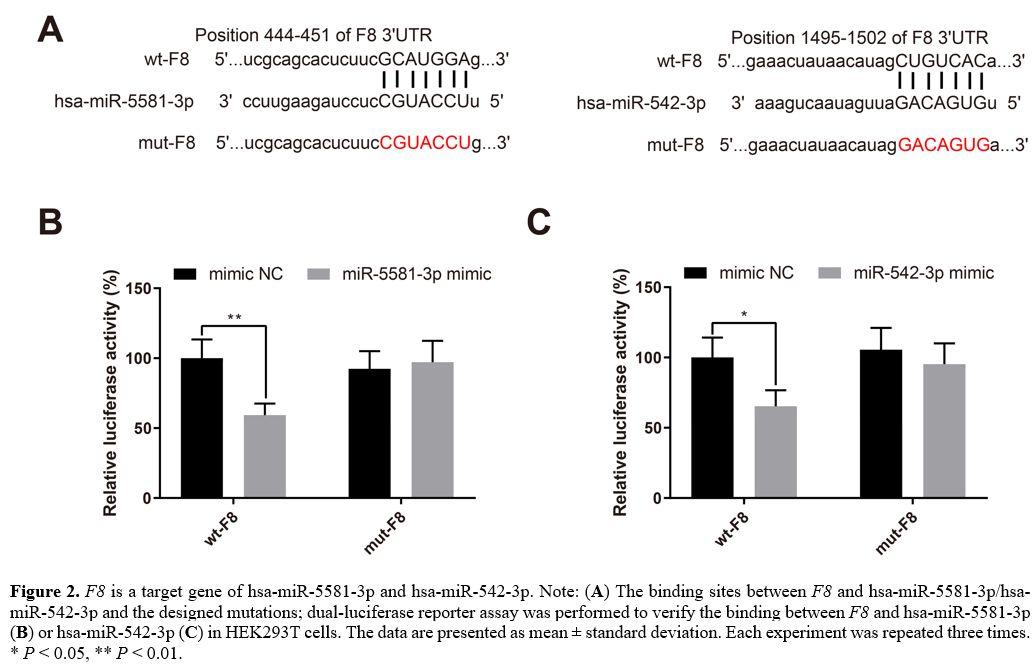 |
Figure
2. F8 is a
target gene of hsa-miR-5581-3p and hsa-miR-542-3p. Note: (A) The binding sites
between F8
and hsa-miR-5581-3p/hsa-miR-542-3p and the designed mutations;
dual-luciferase reporter assay was performed to verify the binding
between F8
and hsa-miR-5581-3p (B)
or hsa-miR-542-3p (C)
in HEK293T cells. The data are presented as mean ± standard deviation.
Each experiment was repeated three times. * P < 0.05, **
P
< 0.01.
|
Hsa-miR-5581-3p
suppresses the expressions of F8 gene and FVIII protein.
HUVECs exhibiting endogenous expression of FVIII were transfected with
mimic NC, hsa-miR-5581-3p mimic, inhibitor NC or hsa-miR-5581-3p
inhibitor. The results of qRT-PCR experiments showed that
hsa-miR-5581-3p was upregulated in the miR-5581-3p mimic group while
downregulated in the miR-5581-3p inhibitor group (Figure 3A, P <
0.001, vs. the mimic NC or inhibitor NC group). The expression of F8 mRNA was
decreased in the miR-5581-3p mimic group while increased in the
miR-5581-3p inhibitor group (Figure
3B, P
< 0.05, vs. the mimic NC or inhibitor NC group). Western blot
analysis showed that the expression of FVIII protein was also decreased
in the miR-5581-3p mimic group while increased in the miR-5581-3p
inhibitor group (Figure 3C, P < 0.05,
vs. the mimic NC or inhibitor NC group). Taken together,
hsa-miR-5581-3p suppresses the expressions of F8 gene and FVIII
protein in HUVECs.
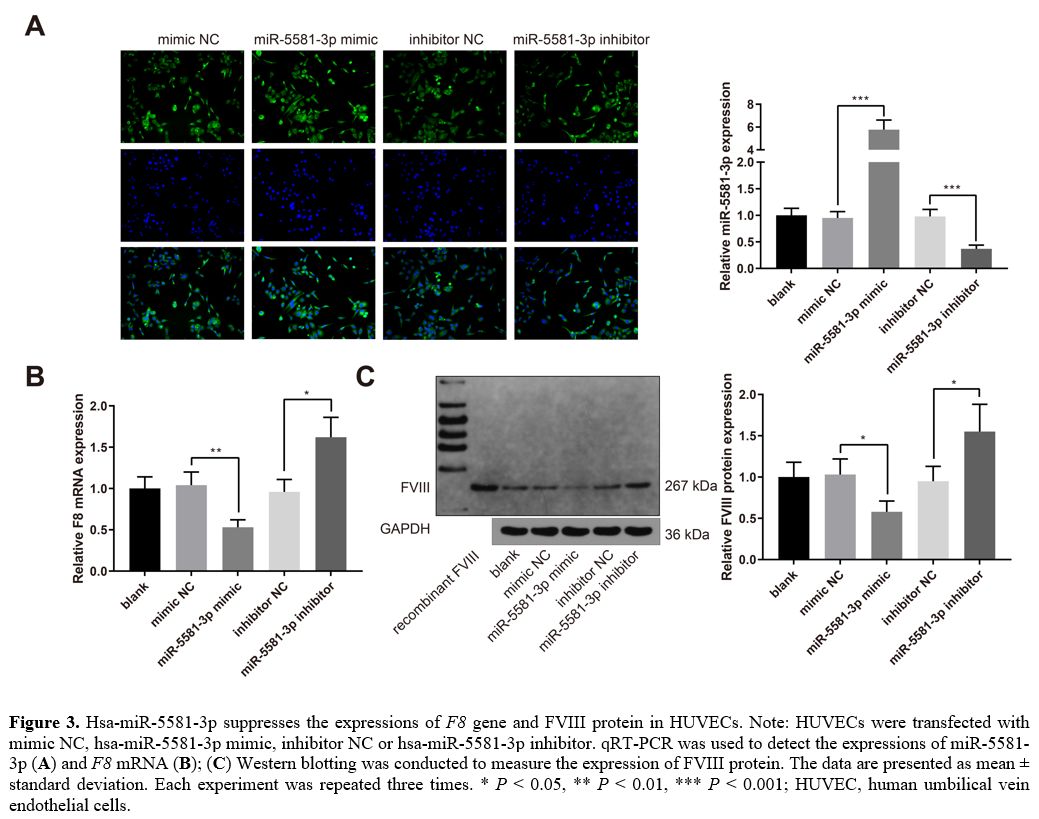 |
Figure
3. Hsa-miR-5581-3p suppresses the expressions of F8
gene and FVIII protein in HUVECs. Note: HUVECs were transfected with
mimic NC, hsa-miR-5581-3p mimic, inhibitor NC or hsa-miR-5581-3p
inhibitor. qRT-PCR was used to detect the expressions of miR-5581-3p (A) and F8 mRNA (B); (C)
Western blotting was conducted to measure the expression of FVIII
protein. The data are presented as mean ± standard deviation. Each
experiment was repeated three times. * P < 0.05, **
P
< 0.01, *** P
< 0.001; HUVEC, human umbilical vein endothelial cells.
|
Hsa-miR-542-3p
inhibits the expressions of F8 gene and FVIII protein.
LCL cells were transfected with mimic NC, hsa-miR-542-3p mimic,
inhibitor NC, or hsa-miR-542-3p inhibitor. The qRT-PCR experiments
indicated that hsa-miR-542-3p was upregulated in the miR-542-3p mimic
group while downregulated in the miR-542-3p inhibitor group (Figure 4A, P < 0.001,
vs. the mimic NC or inhibitor NC group). The expression of F8 mRNA was
decreased in the miR-542-3p mimic group while increased in the
miR-542-3p inhibitor group (Figure
4B, P
< 0.01, vs. the mimic NC or inhibitor NC group). Western blot
analysis showed that the expression of FVIII protein was also decreased
in the miR-542-3p mimic group while increased in the miR-542-3p
inhibitor group (Figure 4C,
P <
0.05, vs. the mimic NC or inhibitor NC group). Taken together,
hsa-miR-542-3p suppresses the expressions of F8 gene and FVIII
protein in LCL cells.
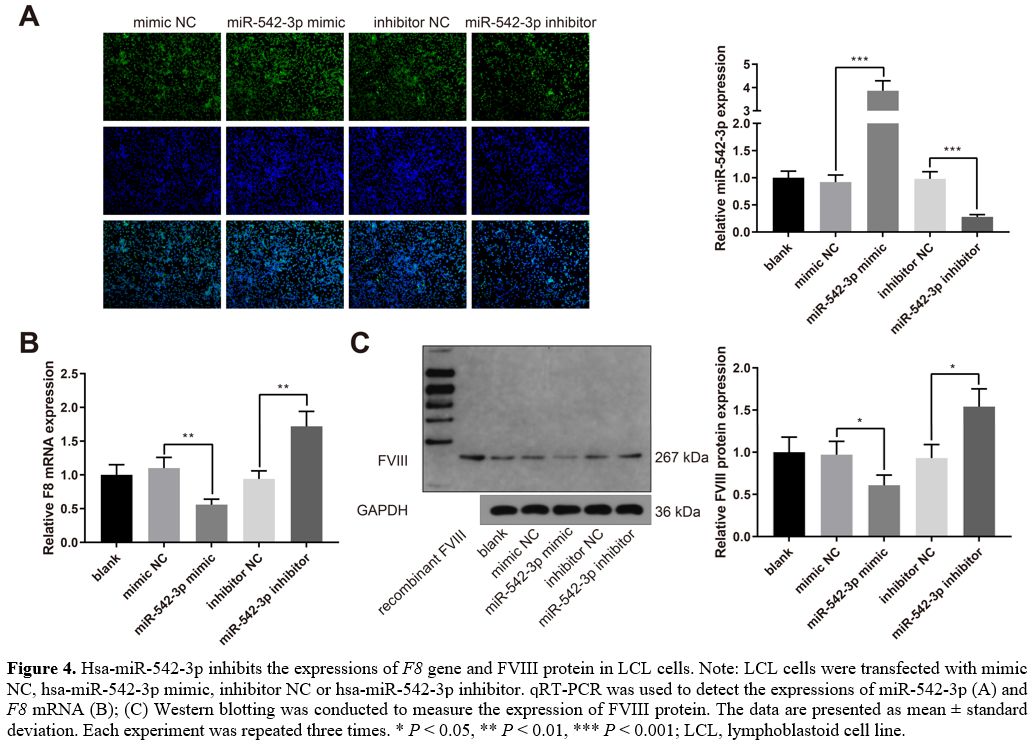 |
Figure
4. Hsa-miR-542-3p inhibits the expressions of F8
gene and FVIII protein in LCL cells. Note: LCL cells were transfected
with mimic NC, hsa-miR-542-3p mimic, inhibitor NC or hsa-miR-542-3p
inhibitor. qRT-PCR was used to detect the expressions of miR-542-3p (A) and F8 mRNA (B); (C)
Western blotting was conducted to measure the expression of FVIII
protein. The data are presented as mean ± standard deviation. Each
experiment was repeated three times. * P < 0.05, **
P
< 0.01, *** P
< 0.001; LCL, lymphoblastoid cell line.
|
Discussion
Numerous
causative DNA variants of the F8
and F9
genes are reported to impact bleeding severity and inhibitor
development in hemophilia patients.[14]
However, no causal mutations in the F8 gene are
detected in a small group of HA patients. Mutations in the 3’UTR of F8 affect the
splicing and expression of mRNA, which leads to reduced FVIII level and
a mild HA.[15]
MiRNAs inhibit their targets through base-pairing interactions with the
3’UTR of target mRNAs, resulting in translation inhibition or mRNA
degradation.[16] In this study,
hsa-miR-5581-3p and hsa-miR-542-3p were found to interact with the F8 gene and
regulate the expression of FVIII.
A total of three databases, including TargetScan, miRDB, and starBase,
were utilized to predict miRNAs that target the 3’UTR of F8. At least two
databases predicted 42 miRNAs. Subsequently, their expressions in the
blood of HA patients with normal F8
genotypes were quantified. Hsa-miR-5581-3p, hsa-miR-30c-3p,
hsa-miR-374b-3p, hsa-miR-542-3p, and hsa-miR-6803-3p were highly
expressed in HA patients. Hsa-miR-5581-3p and hsa-miR-542-3p were
selected for further investigations since they were not reported in
previous studies of HA. In addition, luciferase reporter plasmids were
used to validate the binding sites between
hsa-miR-5581-3p/hsa-miR-542-3p and F8.
The experiment results suggest that F8
is a target of hsa-miR-5581-3p and hsa-miR-542-3p. Furthermore, the
function of these two miRNAs was investigated in HUVECs and LCL that
displayed endogenous expression of FVIII.
Hsa-miR-542-3p suppressed the expressions of F8
mRNA and FVIII in LCL cells. MiR-542-3p is a versatile ncRNA that has
been largely investigated. MiR-542-3p acted as a tumor suppressor in
many cancers such as colorectal cancer,[17]
epithelial ovarian cancer,[18]
hepatocellular carcinoma,[19] and
breast cancer.[20] MiR-542-3p
could also reduce chemoresistance in tumor cells and improve the
antitumor efficacy of chemotherapy.[21]
MiR-542-3p was upregulated in activated hepatic stellate cells and
promoted fibrosis via inhibiting BMP-7.[22]
In kidney, it contributed to fibrosis by targeting AGO1.[23]
Through inhibition of BMP-7, miR-542-3p also suppressed the osteogenic
differentiation of vascular smooth muscle cells in aging rats.[24]
A study by Zhang et al. demonstrated that miR-542-3p facilitated bone
formation by promoting SFRP1-mediated osteogenic differentiation in
mesenchymal stem cells.[25] This
study firstly proved the involvement of hsa-miR-542-3p in HA without F8 mutations.
Overexpression of the-miR-5581-3p decreased the expressions of F8 mRNA and FVIII
in HUVECs, whereas its inhibition increased their expressions.
MiR-5581-3p was the only one F8-targeting
miRNA predicted by all the three databases; however, it has been seldom
reported for its function. Available data showed that overexpression of
miR-5581-3p promoted cellular activities in hepatocellular carcinoma by
inhibiting cardiolipin synthase 1.[26]
This study supplemented the knowledge regarding the physiological
function of hsa-miR-5581-3p.
Conclusions
Hsa-miR-542-3p
and hsa-miR-5581-3p promote HA by targeting F8 mRNA and
inhibiting the expression of FVIII. Not all HA cases are caused by
mutations in the F8
gene, and the pathogenic mechanisms for atypical HA are worthy of
investigation. Emerging evidence has shown that miRNAs can target the
3’UTR of F8
gene and
therefore mediate the expression of FVIII. This study first identifies
the involvement of two novel targets-hsa-miR-542-3p and hsa-miR-5581-3p
in the regulation of FVIII protein, and provides a theoretical basis
for targeting miRNAs in the treatment of HA without F8 mutations.
References (già
inserite)
- Iorio A, Stonebraker JS,
Chambost H, Makris M,
Coffin D, Herr C et al. Establishing the Prevalence and Prevalence at
Birth of Hemophilia in Males: A Meta-analytic Approach Using National
Registries. Ann Intern Med. 2019; 171: 540-546. https://doi.org/10.7326/M19-1208
PMid:31499529
- Peyvandi
F, Garagiola I, Young G. The past and future of haemophilia: diagnosis,
treatments, and its complications. Lancet. 2016; 388: 187-197. https://doi.org/10.1016/S0140-6736(15)01123-X
- Miesbach W, Schwable J,
Muller MM, Seifried E. Treatment Options in Hemophilia. Dtsch Arztebl
Int. 2019; 116: 791-798. https://doi.org/10.3238/arztebl.2019.0791
PMid:31847949 PMCid:PMC6937545
- Nathwani AC. Gene
therapy for hemophilia. Hematology Am Soc Hematol Educ Program. 2019;
2019: 1-8. https://doi.org/10.1182/hematology.2019000007
PMid:31808868 PMCid:PMC6913446
- Garagiola
I, Seregni S, Mortarino M, Mancuso ME, Fasulo MR, Notarangelo LD et al.
A recurrent F8 mutation (c.6046C>T) causing hemophilia A in 8%
of
northern Italian patients: evidence for a founder effect. Mol Genet
Genomic Med. 2016; 4: 152-159. https://doi.org/10.1002/mgg3.189
PMid:27066508 PMCid:PMC4799873
- Donadon
I, McVey JH, Garagiola I, Branchini A, Mortarino M, Peyvandi F et al.
Clustered F8 missense mutations cause hemophilia A by combined
alteration of splicing and protein biosynthesis and activity.
Haematologica. 2018; 103: 344-350. https://doi.org/10.3324/haematol.2017.178327
PMid:29170251 PMCid:PMC5792279
- Johnsen
JM, Fletcher SN, Huston H, Roberge S, Martin BK, Kircher M et al. Novel
approach to genetic analysis and results in 3000 hemophilia patients
enrolled in the My Life, Our Future initiative. Blood Adv. 2017; 1:
824-834. https://doi.org/10.1182/bloodadvances.2016002923
PMid:29296726 PMCid:PMC5727804
- Catalanotto
C, Cogoni C, Zardo G. MicroRNA in Control of Gene Expression: An
Overview of Nuclear Functions. Int J Mol Sci. 2016; 17. https://doi.org/10.3390/ijms17101712
PMid:27754357 PMCid:PMC5085744
- Mohr AM, Mott JL.
Overview of microRNA biology. Semin Liver Dis. 2015; 35: 3-11. https://doi.org/10.1055/s-0034-1397344
PMid:25632930 PMCid:PMC4797991
- Rosset
C, Vieira IA, Salzano FM, Bandinelli E. A germline variant affects
putative miRNA-binding sites at the F8 3'UTR and acts as a potential
haemophilia A phenotype modifier in Southern Brazilian patients.
Haemophilia. 2016; 22: e327-329. https://doi.org/10.1111/hae.12953
PMid:27228178
- Sarachana
T, Dahiya N, Simhadri VL, Pandey GS, Saini S, Guelcher C et al. Small
ncRNA Expression-Profiling of Blood from Hemophilia A Patients
Identifies miR-1246 as a Potential Regulator of Factor 8 Gene. PLoS
One. 2015; 10: e0132433. https://doi.org/10.1371/journal.pone.0132433
PMid:26176629 PMCid:PMC4503767
- Jankowska
KI, McGill J, Pezeshkpoor B, Oldenburg J, Atreya CD, Sauna ZE. Clinical
manifestation of hemophilia A in the absence of mutations in the F8
gene that encodes FVIII: role of microRNAs. Transfusion. 2020; 60:
401-413. https://doi.org/10.1111/trf.15605
PMid:31785023
- Jankowska
KI, Chattopadhyay M, Sauna ZE, Atreya CD. A Foundational Study for
Normal F8-Containing Mouse Models for the miRNA Regulation of
Hemophilia A: Identification and Analysis of Mouse miRNAs that
Downregulate the Murine F8 Gene. Int J Mol Sci. 2020; 21. https://doi.org/10.3390/ijms21165621
PMid:32781510 PMCid:PMC7460574
- Konkle
BA, Johnsen JM, Wheeler M, Watson C, Skinner M, Pierce GF et al.
Genotypes, phenotypes and whole genome sequence: Approaches from the My
Life Our Future haemophilia project. Haemophilia. 2018; 24 Suppl 6:
87-94. https://doi.org/10.1111/hae.13506
PMid:29878652 PMCid:PMC6258054
- Pezeshkpoor
B, Berkemeier AC, Czogalla KJ, Oldenburg J, El-Maarri O. Evidence of
pathogenicity of a mutation in 3' untranslated region causing mild
haemophilia A. Haemophilia. 2016; 22: 598-603. https://doi.org/10.1111/hae.12923
PMid:27216882
- Simonson B, Das S.
MicroRNA Therapeutics: the Next Magic Bullet? Mini Rev Med Chem. 2015;
15: 467-474. https://doi.org/10.2174/1389557515666150324123208
PMid:25807941 PMCid:PMC4410078
- Yuan
L, Yuan P, Yuan H, Wang Z, Run Z, Chen G et al. miR-542-3p inhibits
colorectal cancer cell proliferation, migration and invasion by
targeting OTUB1. Am J Cancer Res. 2017; 7: 159-172.
- Li
J, Shao W, Feng H. MiR-542-3p, a microRNA targeting CDK14, suppresses
cell proliferation, invasiveness, and tumorigenesis of epithelial
ovarian cancer. Biomed Pharmacother. 2019; 110: 850-856. https://doi.org/10.1016/j.biopha.2018.11.104
PMid:30557834
- Tao
J, Liu Z, Wang Y, Wang L, Yao B, Li Q et al. MiR-542-3p inhibits
metastasis and epithelial-mesenchymal transition of hepatocellular
carcinoma by targeting UBE3C. Biomed Pharmacother. 2017; 93: 420-428. https://doi.org/10.1016/j.biopha.2017.06.070
PMid:28666208
- Wu
HX, Wang GM, Lu X, Zhang L. miR-542-3p targets sphingosine-1-phosphate
receptor 1 and regulates cell proliferation and invasion of breast
cancer cells. Eur Rev Med Pharmacol Sci. 2017; 21: 108-114.
- Lyu
H, Wang S, Huang J, Wang B, He Z, Liu B. Survivin-targeting miR-542-3p
overcomes HER3 signaling-induced chemoresistance and enhances the
antitumor activity of paclitaxel against HER2-overexpressing breast
cancer. Cancer Lett. 2018; 420: 97-108. https://doi.org/10.1016/j.canlet.2018.01.065
PMid:29409974 PMCid:PMC6089084
- Ji
F, Wang K, Zhang Y, Mao XL, Huang Q, Wang J et al. MiR-542-3p controls
hepatic stellate cell activation and fibrosis via targeting BMP-7. J
Cell Biochem. 2019; 120: 4573-4581. https://doi.org/10.1002/jcb.27746
PMid:30368874
- Tao
L, Liu X, Da W, Tao Z, Zhu Y. Pycnogenol achieves neuroprotective
effects in rats with spinal cord injury by stabilizing the
mitochondrial membrane potential. Neurol Res. 2020; 42: 597-604. https://doi.org/10.1080/01616412.2020.1773610
PMid:32497471
- Liu
H, Wang H, Yang S, Qian D. Downregulation of miR-542-3p promotes
osteogenic transition of vascular smooth muscle cells in the aging rat
by targeting BMP7. Hum Genomics. 2019; 13: 67. https://doi.org/10.1186/s40246-019-0245-z
PMid:31829291 PMCid:PMC6907335
- Zhang
X, Zhu Y, Zhang C, Liu J, Sun T, Li D et al. miR-542-3p prevents
ovariectomy-induced osteoporosis in rats via targeting SFRP1. J Cell
Physiol. 2018; 233: 6798-6806. https://doi.org/10.1002/jcp.26430
PMid:29319176 PMCid:PMC6001432
- Yin
J, Liu Q, Chen C, Liu W. Small regulatory polypeptide of amino acid
response negatively relates to poor prognosis and controls
hepatocellular carcinoma progression via regulating
microRNA-5581-3p/human cardiolipin synthase 1. J Cell Physiol. 2019;
234: 17589-17599. https://doi.org/10.1002/jcp.28383
PMid:30825207
Supplementary Files
 |
Supplementary
table 1. The 42 F8-targeting
miRNAs predicted by at least two of the bioinformatics tools.
|
[TOP]





