Cong Lu1, Chao Song2, Qingkun Han1 and Xuebing Jing1.
1 Department of Hematology, Zibo Central Hospital, Zibo City, 255000, PR. China.
2 Department of Pathology, Zibo Central Hospital, Zibo City, 255000, PR. China.
Published: July 1, 2022
Received: January 4, 2022
Accepted: June 5, 2022
Mediterr J Hematol Infect Dis 2022, 14(1): e2022047 DOI
10.4084/MJHID.2022.047
This is an Open Access article distributed
under the terms of the Creative Commons Attribution License
(https://creativecommons.org/licenses/by-nc/4.0),
which permits unrestricted use, distribution, and reproduction in any
medium, provided the original work is properly cited.
|
|
Abstract
Background:
It was reported that circular RNA (circRNA) circCTNNA1 plays an
oncogenic role in colorectal cancer, while its role in mantle cell
lymphoma (MCL) is unknown. This study aimed to explore the role of
circCTNNA1 in MCL.
Methods:
Samples of B lymphocytes were collected from 56 MCL patients and 56
healthy controls. The expression of circCTNNA1 and miR-34a in these
samples were determined by RT-qPCR. The direct interaction between
circCTNNA1 and miR-34a in MCL cells was detected using RNA-RNA pulldown
assay. Overexpression assays were performed to study the interactions
between circCTNNA1 and miR-34a. Cell proliferation was assessed with
BrdU assay.
Results: The
results showed that circCTNNA1 was upregulated in MCL and high
expression levels of circCTNNA1 predicted the poor survival of MCL
patients. MiR-34a was downregulated in MCL and inversely correlated
with circCTNNA1. CircCTNNA1 was predicted to interact with miR-34a, and
the interaction between them was confirmed by RNA pull-down assay.
Interestingly, overexpression of circCTNNA1 and miR-34a did not affect
the expression of each other. Cell proliferation analysis showed that
overexpression of circCTNNA1 reversed the inhibitory effects of
overexpression of miR-34a on cell proliferation.
Conclusion:
Upregulation of circCTNNA1 in MCL predicts poor survival of patients
and it may sponge miR-34a to promote cancer cell proliferation.
|
Introduction
As
a cancer develops in B-lymphocytes, mantle cell lymphoma (MCL) is a
rare type of non-Hodgkin lymphoma that mainly affects patients older
than 60 years old, and it is more common in men than that in women. [1-3]
MCL affects about 1 case per 200, 000 people annually. In China, about
4.2 out of 100,000 people will develop MCL during their life time and
more than half of these patients will die of MCL.[4]
Although the clinical application of rituximab and novel agents, as
well as the utilization of hematopoietic cell transplantation in the
treatment of MCL have significantly increased the survival of patients,
there is still no cure for MCL.[5-7] Actually, no more
than 50% of MCL patients can survive more than 5 years after the
initial diagnosis, and the 10-year overall survival rate is below 10%.[7] Therefore, more effective approaches are still needed to improve the outcomes of MCL treatment.
The development of novel anti-MCL approaches needs better elucidation of molecular factors involved in this malignancy.[7-11]
Recent studies have identified several factors, such as the NF-κB
signal pathway and CDK7, play critical roles in MCL and might serve as
potential therapeutic targets for the treatment of MCL.[9,11]
Circular RNAs (circRNAs) are emerging novel players in human cancers
that regulate cancer progression mainly by affecting gene expression.[12,13]
Therefore, circRNAs are promising targets to treat cancers including
MCL. CircRNA circCTNNA1 plays an oncogenic role in colorectal cancer,[14]
while its role in MCL is unknown. We predicted that circCTNNA1 could
interact with miR-34a, which plays critical roles in other types of
B-cell lymphoma.[15] We therefore speculated that
circCTNNA1 could interact with miR-34a to participate in MCL. This
study was carried out to investigate the interaction between circCTNNA1
and miR-34a in MCL.
Materials and Methods
MCL patients and survival analysis.
This study enrolled a total of 56 MCL patients (34 cases of blastoid
variant and 22 cases of pleomorphic variant; 63.4+/-6,6 years; 32 males
and 24 females) and 56 healthy controls (63.5+/-6,5 years; 32 males and
24 females) at Zibo Central Hospital from May 2014 to May 2016. Ethics
approval was obtained from the Ethics Committee of this hospital. Two
groups of participants showed similar distributions of age and gender.
The diagnosis of MCL was performed according to WHO classification of
hematological neoplasms. Patients with leukemic phase MCL were excluded
from this study. Fasting blood samples were collected from both groups
of participants prior to the initiation of therapies. DETACHaBEAD CD19
Kit (Invitrogen) was used to separate B lymphocytes from all blood
samples. Purity of higher than 90% was reached in all samples of B
lymphocytes. All patients signed the informed consent. MCL patients
were treated with chemoimmunotherapy R-CHOP, in which rituximab,
cyclophosphamide, hydroxydaunomycin, vincristine sulfate and prednisone
were included.
All 56 MCL patients were followed up for 5 years to
evaluate the prognostic value of circCTNNA1 for MCL. Patients were
visited through outpatient visit and telephone in some cases. All
patients either died of MCL or survived until the end of follow-up.
This study was conducted in accordance with the Declaration of Helsinki.
Cells and transfections.
Two human MCL cell lines JVM-2 and Z138 were obtained from the Cell
Bank of Chinese Academy of Sciences (Shanghai, China). Cells were
cultivated in DMEM (GIBCO) containing FBS (10%), penicillin (50
units/mL) and streptomycin (50 mg/mL) in an incubator with temperature,
humidity and CO2 set to 37 °C, 95% and 5%, respectively.
Overexpression
of circCTNNA1 and miR-34a was achieved in cells using Neon
Electroporation Transfection (Thermo Fisher Scientific) to transfect
circCTNNA1-pcDNA3.1 vector or miR-34a mimic. The dosages of vector and
miRNA were 50 and 150 µM for 107 cells, respectively. The amount of
vector and miRNA was blow 10% of the total volume.
RNA preparations.
RNAstorm™ RNA Isolation Kit (Biotium) was used to extract total RNAs
from both cultivated cells and tissue samples. DNase treatment was also
included in this kit. All steps of RNA isolation and purification were
carried out following the manufacturer’s instructions. RNA
concentrations were measured and RNA integrity was analyzed with
Bioanalyzer. RNA samples with a RNA integrity number (RIN) higher than
8.5 were used in subsequent assays.
Reverse transcriptions and qPCR.
Reverse transcriptions (RTs) were performed to prepare cDNA samples
with 3 µg total RNAs as template using SSRT IV (Invitrogen). Briefly, a
13 µl mixture of RNA samples, primer (0.5 µl) and 10 mM dNTP (1 µl)
were was incubated at 65 °C for 5 min, followed by incubation on ice
for 2 min. After that, 1 µl RNaseout, 1 µl DTT, 1 µl reverse
transcriptase and 4 µl 5x buffer were added to prepare a 20 µl mixture,
which was then incubated at 23 °C for 5 min, 50 °C for 15 min and 80 °C
for 10 min. cDNA samples were used as template to perform qPCRs (1 µl
cDNA for 20 µl reaction system). The expression of circCTNNA1 and
miR-34a were determined with 18S rRNA and U6 as the internal control,
respectively. The 2−ΔΔCt method was used to normalize Ct values.
RNA-RNA pulldown assay. A circCTNNA1 expression vector with T7 promoter was used to prepare circCTNNA1 and NC RNA in vitro
transcripts using T7 reverse transcriptase (NEB). Both transcripts were
labeled with biotin using Pierce™ RNA 3’ End Biotinylation Kit (Thermo
Fisher Scientific). The labeled RNAs were purified and named Bio-
circCTNNA1 and Bio-NC, respectively. Neon Electroporation Transfection
was used to transfect both labeled RNAs into cells, followed by cell
culture for 48 h. After that, cell lysis was prepared to isolate RNA
complex with magnetic beads. RNA samples were purified and quantified.
RT-qPCR was then performed to quantify the expression levels of
miR-34a.
BrdU incorporation cell proliferation assay.
BrdU measurement was performed to analyze DNA synthesis, which directly
reflects cell proliferation. Cell culture was performed in 96-well cell
culture plates with 3 replicate wells set for each experiment. Cell
culture was performed for 48 h, then 10 μM BrdU (Sigma-Aldrich) was
added to culture medium, followed by incubation for 2 h. After
incubation, peroxidase-coupled anti-BrdU-antibody (Sigma-Aldrich) was
added, then peroxidase substrate (tetramethylbenzidine) was added at 60
min later.
Statistical analysis.
Mean ± standard deviation (SD) values of at least 3 biological
replicates were presented. Shapiro-Wilk test was applied to test data
distribution. Mann-Whitney test was used to explore differences with
statistical significance between groups using GraphPad Prism software
(GraphPad). The 56 patients were divided into high and low circCTNNA1
level groups (n = 28, median expression level of circCTNNA1 as the
cutoff value). Survival curves were plotted and log-rank test was used
to compare curves. Associations between patients’ clinical features and
the expression of circCTNNA1 were analyzed with Chi-squared test. Cox’s
proportional hazards regression was used for univariate and
multivariate analysis. p < 0.05 was considered as statistically significant.
Results
The expression of circCTNtNA1 in MCL and its prognostic value for this malignancy.
The expression of circCTNNA1 in B lymphocytes samples of MCL patients
and healthy controls were detected by performing RT-qPCRs. The results
showed that the expression levels of circCTNNA1 were significantly
increased in MCL patients (Figure 1A, p
< 0.01). Survival analysis showed that the poor survival of MCL
patients was closely correlated with patients’ high expression levels
of circCTNNA1 (Figure 1B). No
obvious differences in treatment methods and drug dosages were observed
between high and low level circCTNNA1 groups. It is worth noting that
similar results were obtained using Youden index as cutoff value for
grouping (data not shown). Chi-squared test showed that the expression
levels of circCTNNA1 were not closely correlated with patients’ age,
gender, WBC, clinical stages, ECOG and serum LDH (Table 1).
Univariate and multivariate analysis showed that circCTNNA1 was a
significant prognostic factor for the overall survival of MCL patients (Table 2).
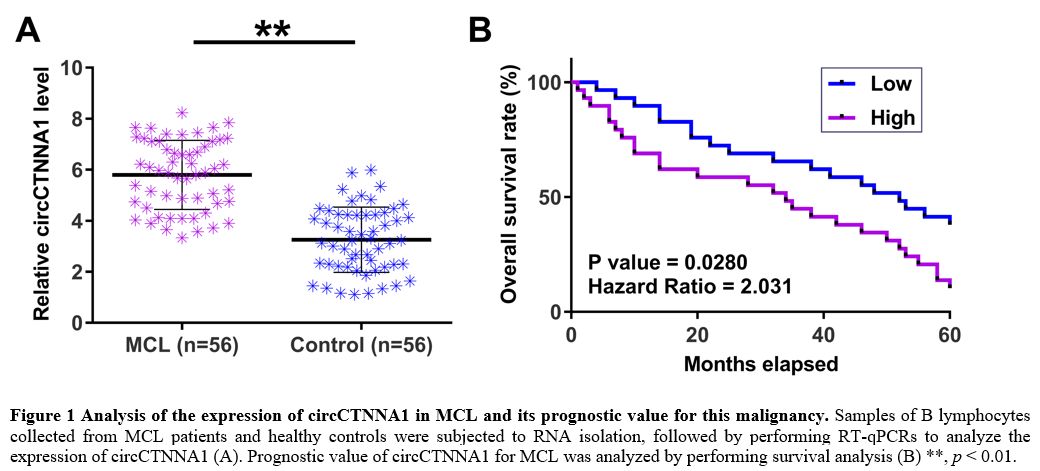 |
Figure 1. Analysis of the expression of circCTNNA1 in MCL and its prognostic value for this malignancy.
Samples of B lymphocytes collected from MCL patients and healthy
controls were subjected to RNA isolation, followed by performing
RT-qPCRs to analyze the expression of circCTNNA1 (A). Prognostic value
of circCTNNA1 for MCL was analyzed by performing survival analysis (B)
**, p < 0.01. |
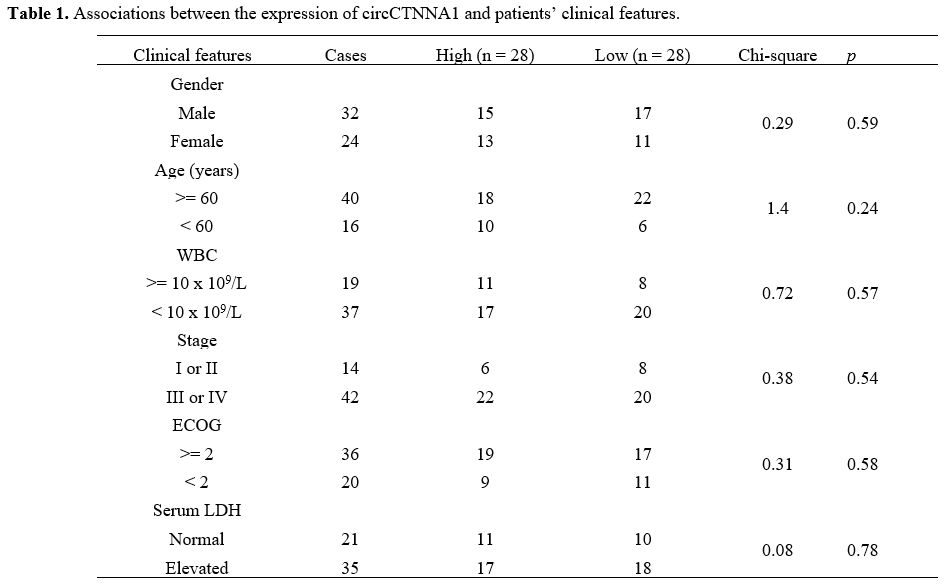 |
Table
1. Associations between the expression of circCTNNA1 and patients’ clinical features. |
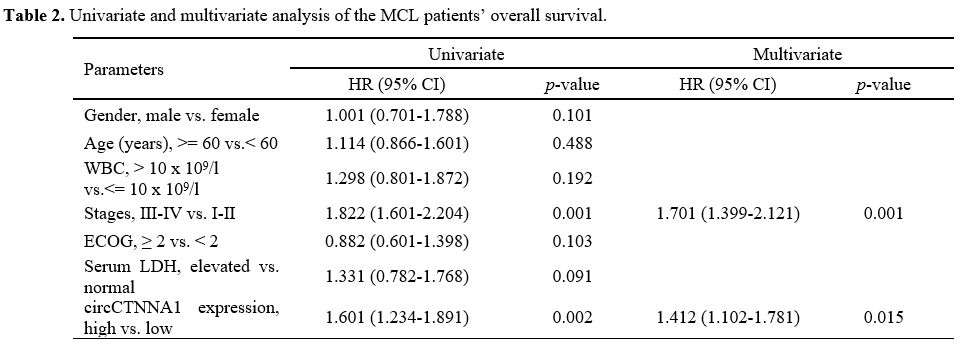 |
Table
2. Univariate and multivariate analysis of the MCL patients’ overall survival. |
The expression of miR-34a in MCL and its correlations with circCTNNA1.
The expression of miR-34a in B lymphocytes samples of MCL patients and
healthy controls were also detected by performing RT-qPCRs. The results
revealed significantly downregulated miR-34a in MCL compared to that in
the Control group (Figure 2A, p
< 0.01). Pearson’s correlation coefficient showed that the
expression of miR-34a was inversely correlated with circCTNNA1 across
both MCL samples (Figure 2B) and the Control samples (Figure 2C).
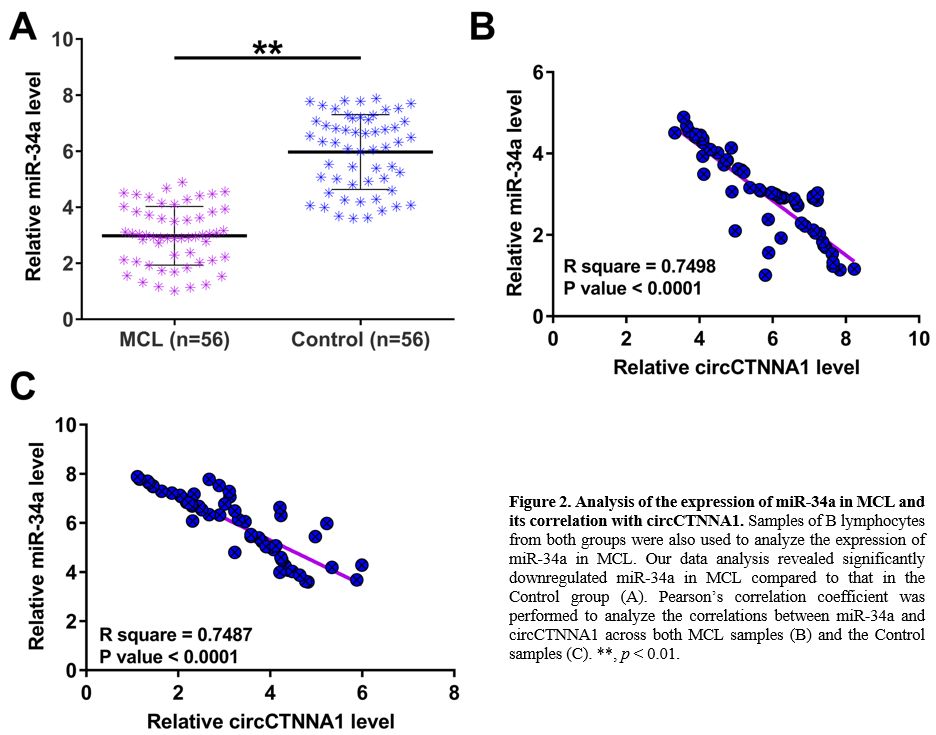 |
Figure
2. Analysis of the expression of miR-34a in MCL and its correlation with circCTNNA1.
Samples of B lymphocytes from both groups were also used to analyze the
expression of miR-34a in MCL. Our data analysis revealed significantly
downregulated miR-34a in MCL compared to that in the Control group (A).
Pearson’s correlation coefficient was performed to analyze the
correlations between miR-34a and circCTNNA1 across both MCL samples (B)
and the Control samples (C). **, p < 0.01. |
CircCTNNA1 and miR-34a directly interacted with each other.
The potential base pairing could be formed between circCTNNA1 and
miR-34a was predicted using the online program IntaRNA 2.0. It was
observed that circCTNNA1 and miR-34a could form strong base pairing (Figure 3A).
RNA pull-down assay showed that compared to Bio-NC group,
Bio-circCTNNA1 exhibited significantly higher expression levels of
miR-34a (Figure 3B, p < 0.01). Therefore, circCTNNA1 and miR-34a could directly interact with each other.
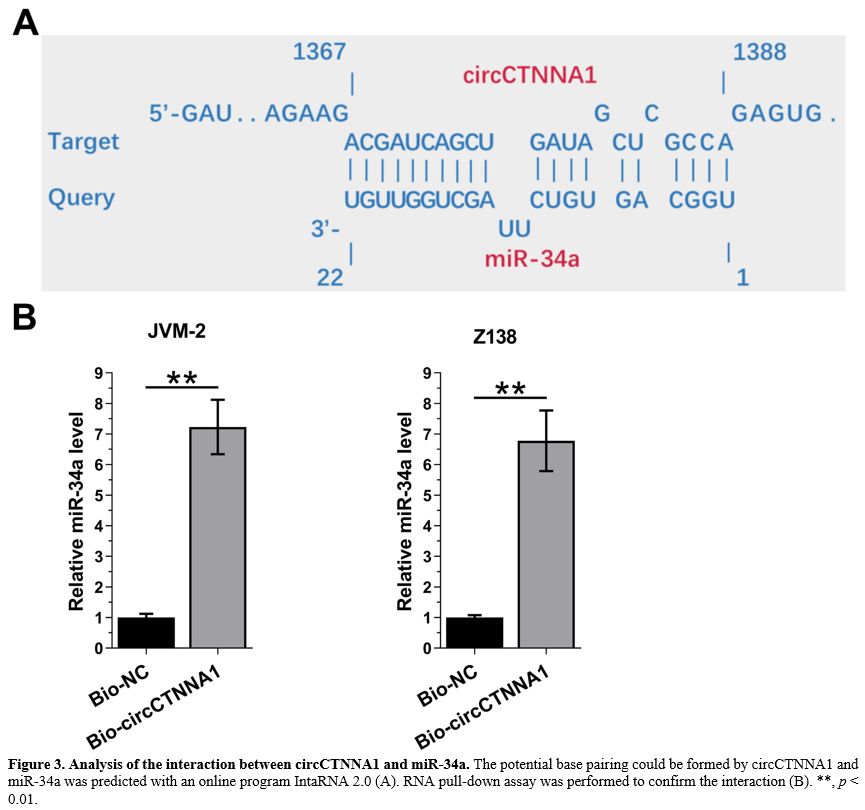 |
Figure 3. Analysis of the interaction between circCTNNA1 and miR-34a.
The potential base pairing could be formed by circCTNNA1 and miR-34a
was predicted with an online program IntaRNA 2.0 (A). RNA pull-down
assay was performed to confirm the interaction (B). **, p < 0.01. |
The role of miR-34a and circCTNNA1 in regulating the expression of each other.
Overexpression of miR-34a or circCTNNA1 were achieved in both JVM-2 and
Z138 cells. It was observed that miR-34a and circCTNNA1 were
significantly overexpressed from 24 h to 96 h (Figure 4A, p < 0.05). Interestingly, miR-34a showed no role in regulating the expression of circCTNNA1 (Figure 4B). Moreover, circCTNNA1 also did not alter the expression of miR-34a (Figure 4C).
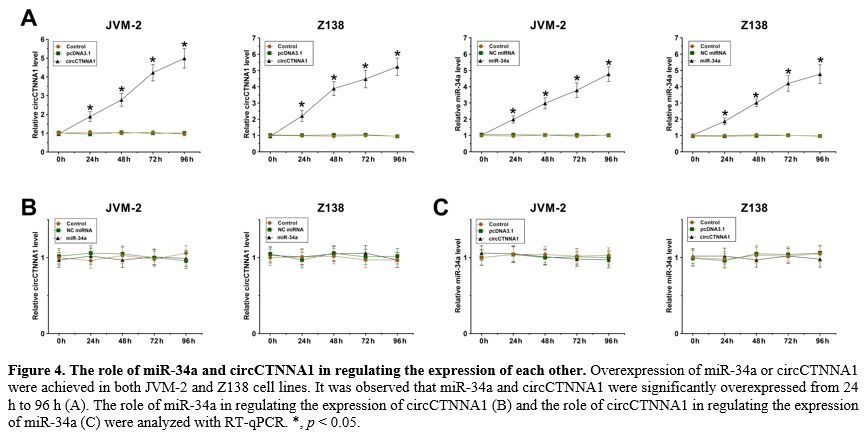 |
Figure
4. The role of miR-34a and circCTNNA1 in regulating the expression of each other.
Overexpression of miR-34a or circCTNNA1 were achieved in both JVM-2 and
Z138 cell lines. It was observed that miR-34a and circCTNNA1 were
significantly overexpressed from 24 h to 96 h (A). The role of miR-34a
in regulating the expression of circCTNNA1 (B) and the role of
circCTNNA1 in regulating the expression of miR-34a (C) were analyzed
with RT-qPCR. *, p < 0.05. |
The role of circCTNNA1 and miR-34a in the proliferation of MCL cells.
BrdU assay was performed to evaluate the proliferation of MCL cells
with overexpression of circCTNNA1 and/or miR-34a. It showed that
miR-34a suppressed cell proliferation, while circCTNNA1 increased cell
proliferation. Moreover, overexpression of circCTNNA1 totally reversed
the inhibitory effects of overexpression of miR-34a on cell
proliferation (Figure 5, p < 0.05).
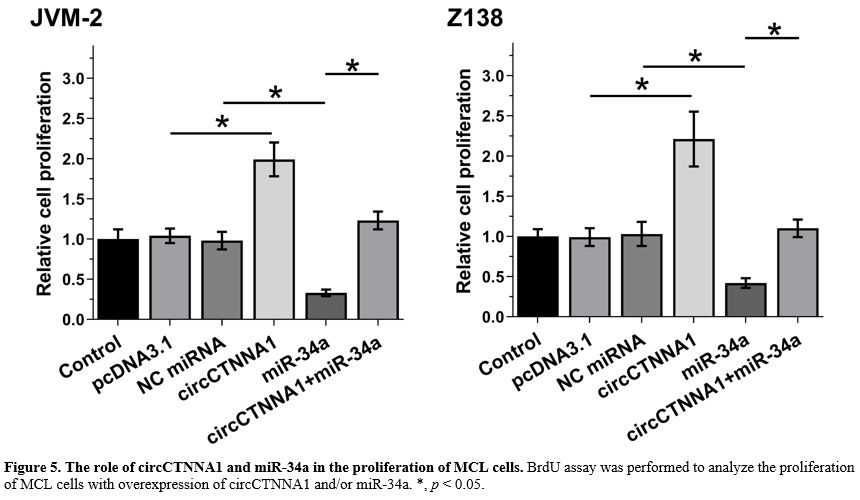 |
Figure
5. The role of circCTNNA1 and miR-34a in the proliferation of MCL cells. BrdU assay was performed to analyze the proliferation of MCL cells with overexpression of circCTNNA1 and/or miR-34a. *, p < 0.05. |
Discussion
The
present study explored the interaction between circCTNNA1 and miR-34a
in MCL. We found that circCTNNA1 was highly expressed in MCL and it
predicted the poor survival of MCL patients. Moreover, circCTNNA1 may
sponge miR-34a to suppress its role in inhibiting MCL cell
proliferation.
It has been well established that circCTNNA1 plays oncogenic roles in colorectal cancers.[14,16]
Two previous studies showed that circCTNNA1 sponges miR-149-5p and
miR-363-3p to upregulate FOXM1 and CXCL5, respectively, to promote the
progression of MCL.[14,16] However,
the prognostic value of circCTNNA1 for human cancers remains unclear.
The present study revealed upregulation of circCTNNA1 in MCL. In
addition, overexpression of circCTNNA1 significantly increased the
proliferation of MCL cells. Therefore, circCTNNA1 is likely an
oncogenic circRNA in MCL. Moreover, our survival analysis showed that
the high expression levels of circCTNNA1 were closely correlated with
poor survival of MCL patients. Interestingly, the expression levels of
circCTNNA1 in MCL patients were not correlated with their clinical
features that may affect survival, such as clinical stages, age and
WBC. Therefore, circCTNNA1 is likely an independent prognostic factor
for MCL.
Although the involvement of miR-34a in MCL is unclear,
its role in other types of B-cell lymphoma, such as diffuse large
B-cell lymphoma, has been reported.[15] MiR-34a was
downregulated in diffuse large B-cell lymphoma and overexpression of
miR-34a increased the sensitivity of cancer cells to doxorubicin,
thereby improving treatment outcomes. This study reported the decreased
expression levels of miR-34a in MCL. In addition, decreased
proliferation of MCL cells was observed after the overexpression of
miR-34a. Therefore, miR-34a is a tumor suppressor in MCL.
Interestingly, circCTNNA1 directly interacted with miR-34a, while they
showed no role in regulating the expression of each other. Moreover,
circCTNNA1 suppressed the role of miR-34a in inhibiting cancer cell
proliferation. Therefore, circCTNNA1 may only sponge miR-34a, but not
regulate its expression in MCL cells to promote cancer cell
proliferation in MCL.
This study characterized a novel
circCTNNA1/miR-34a pathway in MCL. However, expression of circCTNNA1
and the role of circCTNNA1/miR-34a pathway in other types of B-cell
lymphoma is unclear. Future studies may focus on the involvement of
circCTNNA1/miR-34a in other types of B-cell lymphoma.
Conclusions
In
conclusion, circCTNNA1 is likely an oncogenic circRNA in MCL. It may
promote MCL cell proliferation by sponging miR-34a. Moreover, measuring
the expression levels of circCTNNA1 in MCL patients may assist the
prognosis of MCL, thereby improving patients’ survival.
References
- Cheah CY, Seymour JF, Wang ML. Mantle Cell Lymphoma. J Clin Oncol .2016;34:1256-69. https://doi.org/10.1200/JCO.2015.63.5904 PMid:26755518
- Jain
P, Wang M. Mantle cell lymphoma: 2019 update on the diagnosis,
pathogenesis, prognostication, and management. Am J Hematol.
2019;94:710-25. https://doi.org/10.1002/ajh.25487 PMid:30963600
- Cortelazzo S, Ponzoni M, Ferreri AJM, Dreyling M. Mantle cell lymphoma. Crit Rev Oncol Hematol .2020;153:103038. https://doi.org/10.1016/j.critrevonc.2020.103038 PMid:32739830
- Chen
W, Sun K, Zheng R, Zeng H, Zhang S, Xia C, Yang Z, Li H, Zou X, He J.
Cancer incidence and mortality in China, 2014. Chin J Cancer Res.
2018;30:1-12. https://doi.org/10.21147/j.issn.1000-9604.2018.01.01 PMid:29545714 PMCid:PMC5842223
- Maddocks K. Update on mantle cell lymphoma. Blood. 2018;132:1647-56. https://doi.org/10.1182/blood-2018-03-791392 PMid:30154113
- Rule
S, Dreyling M, Goy A, Hess G, Auer R, Kahl B, Hernández-Rivas J, Qi K,
Deshpande S, Parisi L, Wang M. Ibrutinib for the treatment of
relapsed/refractory mantle cell lymphoma: extended 3.5-year follow up
from a pooled analysis. Haematologica. 2019;104:e211-e4. https://doi.org/10.3324/haematol.2018.205229 PMid:30442728 PMCid:PMC6518912
- Kluin-Nelemans
HC, Hoster E, Hermine O, Walewski J, Geisler CH, Trneny M, Stilgenbauer
S, Kaiser F, Doorduijn JK, Salles G, Szymczyk M, Tilly H, Kanz L,
Schmidt C, Feugier P, Thieblemont C, Zijlstra JM, Ribrag V, Klapper W,
Pott C, Unterhalt M, Dreyling MH. Treatment of Older Patients With
Mantle Cell Lymphoma (MCL): Long-Term Follow-Up of the Randomized
European MCL Elderly Trial. J Clin Oncol. 2020;38:248-56. https://doi.org/10.1200/JCO.19.01294 PMid:31804876
- Vogt N, Dai B, Erdmann T, Berdel WE, Lenz G. The molecular pathogenesis of mantle cell lymphoma. Leuk Lymphoma. 2017;58:1530-7. https://doi.org/10.1080/10428194.2016.1248965 PMid:27894215
- Balaji
S, Ahmed M, Lorence E, Yan F, Nomie K, Wang M. NF-κB signaling and its
relevance to the treatment of mantle cell lymphoma. J Hematol Oncol.
2018;11:83. https://doi.org/10.1186/s13045-018-0621-5 PMid:29907126 PMCid:PMC6002979
- Klener P. Advances in Molecular Biology and Targeted Therapy of Mantle Cell Lymphoma. Int J Mol Sci. 2019;20. https://doi.org/10.3390/ijms20184417 PMid:31500350 PMCid:PMC6770169
- Choi
YJ, Kim DH, Yoon DH, Suh C, Choi CM, Lee JC, Hong JY, Rho JK. Efficacy
of the novel CDK7 inhibitor QS1189 in mantle cell lymphoma. Sci Rep
.2019;9:7193. https://doi.org/10.1038/s41598-019-43760-z PMid:31076643 PMCid:PMC6510728
- Shang Q, Yang Z, Jia R, Ge S. The novel roles of circRNAs in human cancer. Mol Cancer. 2019;18:6. https://doi.org/10.1186/s12943-018-0934-6 PMid:30626395 PMCid:PMC6325800
- Lei
B, Tian Z, Fan W, Ni B. Circular RNA: a novel biomarker and therapeutic
target for human cancers. Int J Med Sci. 2019;16:292-301. https://doi.org/10.7150/ijms.28047 PMid:30745810 PMCid:PMC6367529
- Chen
P, Yao Y, Yang N, Gong L, Kong Y, Wu A. Circular RNA circCTNNA1
promotes colorectal cancer progression by sponging miR-149-5p and
regulating FOXM1 expression. Cell Death Dis. 2020;11:557. https://doi.org/10.1038/s41419-020-02757-7 PMid:32699205 PMCid:PMC7376054
- Marques
SC, Ranjbar B, Laursen MB, Falgreen S, Bilgrau AE, Bødker JS, Jørgensen
LK, Primo MN, Schmitz A, Ettrup MS, Johnsen HE, Bøgsted M, Mikkelsen
JG, Dybkær K. High miR-34a expression improves response to doxorubicin
in diffuse large B-cell lymphoma. Exp Hematol. 2016;44:238-46.e2. https://doi.org/10.1016/j.exphem.2015.12.007 PMid:26854484
- Zhang
Y, Zheng S, Liao N, Huang H, Chen W, Wu Z, Wu D. CircCTNNA1 acts as a
ceRNA for miR-363-3p to facilitate the progression of colorectal cancer
by promoting CXCL5 expression. J Biol Res (Thessalon) .2021;28:7. https://doi.org/10.1186/s40709-021-00135-8 PMid:33640021 PMCid:PMC7913448
[TOP]






