Hong Li, Kehong Bi, Saran Feng, Yan Wang and Chuansheng Zhu.
Department of
Hematology, The First Affiliated Hospital of Shandong First Medical
University & Shandong Provincial Qianfoshan Hospital, Shandong
Province, P.R. China.
Correspondence to: Hong
Li, Department of Hematology, The First Affiliated Hospital of Shandong
First Medical University & Shandong Provincial Qianfoshan Hospital,
No. 16766 Jingshi Road, Jinan 250014, Shandong Province, P.R. China
Email:
honglimedical6@163.com
Published: September 1, 2022
Received: April 20, 2022
Accepted: August 7, 2022
Mediterr J Hematol Infect Dis 2022, 14(1): e2022062 DOI
10.4084/MJHID.2022.062
This is an Open Access article distributed
under the terms of the Creative Commons Attribution License
(https://creativecommons.org/licenses/by-nc/4.0),
which permits unrestricted use, distribution, and reproduction in any
medium, provided the original work is properly cited.
|
|
Abstract
Background: MicroRNA-129
(miR-129) is known to promote chemosensitivity of many types of cancer
cells. However, the role of miR-129 in acute myeloid leukaemia (AML) is
unclear. We predicted that premature miR-129 might interact with
circEHBP1, a well-characterized oncogene in bladder cancer, and
analyzed the interaction between circEHBP1 and miR-129 in AML.
Methods:
Expression of circEHBP1 and miR-129 in AML patients before and after
adriamycin (ADR) treatment was determined by RT-qPCR. CircEHBP1
distribution in nuclear and cytoplasm fractions of AML cells was
determined using a cellular fractionation assay. The direct interaction
of circEHBP1 with premature miR-129 was explored with an RNA-RNA
pull-down assay. Finally, the role of circEHBP1 in regulating miR-129
maturation was analyzed in overexpression cells by RT-qPCRs.
Results:
Compared to the controls, AML patients exhibited increased circEHBP1
and premature miR-129 levels but decreased mature miR-129 levels.
Altered gene expression was more obvious in ADR resistant group than in
ADR sensitive group. CircEHBP1 was detected in both nuclear and
cytoplasm fractions of AML cells and directly interacted with premature
miR-129. CircEHBP1 overexpression increased premature miR-129 level but
decreased mature miR-129 level. In AML cells, circEHBP1 suppressed
ADR-induced cell apoptosis and attenuated the enhancing effects of
miR-129 on cell apoptosis. More importantly, the role of circEHBP1 in
regulating cell apoptosis was more obvious in ADR resistance cells.
Conclusion: CircEHBP1 may suppress miR-129 maturation to increase the chemoresistance of cancer cells to ADR in AML.
|
Introduction
As
a type of blood malignancy developing in bone marrow, acute myeloid
leukaemia (AML) originates from blood-forming cells and mainly affects
people older than 45 years old.[1,2] Compared to other types of leukaemia, AML deteriorates quickly without proper treatment.[3,4] Even with appropriate treatment, fewer than 30% of AML patients can survive more than 5 years.[5]
Although some novel therapeutic approaches, such as targeted therapy
and stem cell transplantation, have been developed to treat AML,
chemotherapies, including adriamycin (ADR), remains the most widely
used AML treatment modality.[6-8] However, chemoresistance will inevitably develop in many cases after treatment, leading to a poor prognosis.
Circular RNAs (circRNAs), a novel type of endogenous non-coding RNA (ncRNA), play vital roles in various human cancers.[9] Although abnormally regulated circRNAs in AML, including circMYBL2,[10] circ_0001947,[11] and circ_0079480,[12]
are considered promising biomarkers for the diagnosis and treatment of
AML, the roles of circRNAs in AML chemotherapy resistance are still not
well understood. CircEHBP1 has been reported to promote bladder cancer
progression via the miR-130a-3p/TGFβR1/VEGF-D signaling.[13] Thus, this study extensively explored its functions in AML cells.
MicroRNAs (miRNAs) are a class of ncRNAs with about 22 nucleotides in length.[14]
The expression of miRNAs is frequently altered in AML, and changes in
miRNA expression may trigger AML deterioration or remission.[15]
MiR-129 is known to promote chemosensitivity of cancer cells in many
types of cancers and plays a critical player in developing
chemosensitivity.[16-18] However, its role in AML is
still unclear. Therefore, we predicted that premature miR-129 might
interact with circEHBP1, a characterized oncogene in bladder cancer,[19] and hypothesized that circEHBP1 might also participate in chemosensitivity via interaction with miR-129 in AML.
Materials and Methods
Clinical samples.
The present study included bone marrow (BM) specimens donated by a
total of 60 AML patients (38 males and 22 females, 63.4 ± 6.9 years
old) and 48 bone marrow transplantation donors (29 males and 19
females, 63.7 ± 7.1 years old). All these participants were admitted to
The First Affiliated Hospital of Shandong First Medical University
& Shandong Provincial Qianfoshan Hospital between May 2019 and May
2021. This study was carried out after obtaining approval from the
Ethics Committee of The First Affiliated Hospital of Shandong First
Medical University & Shandong Provincial Qianfoshan Hospital in
compliance with the principles of the most recent version of the
Declaration of Helsinki. AML patients were diagnosed through multiple
approaches, including immune phenotyping and bone marrow routine. Only
AML patients who received ADR treatment were enrolled. The 60 patients
were grouped into ADR-sensitive (n=40) and resistant (n=20) groups
based on treatment outcomes. Patients in the sensitive group were
remitted completely or partially, and patients in the resistant group
were at stable status or underwent deterioration. Bone marrow
mononuclear cells (BMMCs) were isolated using Ficoll-Isopaque Plus
(Sigma-Aldrich) through density gradient centrifugation and stored in a
liquid nitrogen tank prior to the subsequent assays. All participants
signed the written informed consent.
AML cells and cell culture.
ADR-sensitive cell line (HL60) and ADR-resistant cell line (HL60/ADR)
were purchased from BeNa Culture Collection (Suzhou) and cultured in
DMEM (Gibco, USA) supplemented with 10% FBS (Sigma-Aldrich, USA) and 1%
antibiotic-antimycotic mixture (Thermo Fisher Scientific) at 37°C in an
incubator with 5% CO2 and 95% humidity.
Cell transfections.
Both HL60 and HL60/ADR cells were overexpressed with circEHBP1 or
miR-129 through the transfections of circEHBP1 expression vector or
mimic of miR-129 using Lipofectamine 2000 Transfection Reagent (Thermo
Fisher Scientific) following the manufacturer's protocol. In each
transfection, 2x106 cells were
transfected with 10 nM vector or 50 nM miRNA mimicking Lipofectamine
2000 use. After incubation with a transfection mixture for 6h, cells
were further cultured in a fresh medium for 48h, and overexpression was
confirmed by RT-qPCRs.
MTT (3-(4,5-Dimethylthiazol-2-yl) assay.
ADR sensitivity was determined using the IC50 value (half maximal
inhibitory concentration) of ADR. By calculating the cell inhibition
rate of ADR treatment with different concentrations (0, 2, 4, 8, 16,
32, 64, and 128 μM), the representative curve of AML cell growth
inhibition was plotted. ADR concentration corresponding to 50% cell
viability in the inhibitory rate curve was IC50 value.
RNA preparations.
Total RNA was isolated from both BMMCs and in vitro cultured HL60 cells
using EasyPure® RNA Purification Kit (TransGen Biotech Co., LTD) and
eluted in RNase-free water. After removal of contaminated genomic DNA
using DNase I digestion, RNA samples were subjected to Bioanalyzer
analysis to determine their integrity and determination. RNA samples
were re-isolated if their quality was unsatisfactory (RIN value <
8.0 and/or RNA concentration < 500 ng/μl).
RT-qPCR.
About 1000ng total RNA from each sample was reverse transcribed into
cDNA using LunaScript® RT SuperMix Kit (NEB). qPCRs were performed to
determine the expression of circEHBP1 and premature miR-129 using 18S
rRNA as the internal control, and mature miR-129 expression was
analyzed using All-in-One™ miRNA qRT-PCR Reagent Kits (Genecopoeia)
with U6 as the internal control. Ct values were processed using the
method of 2−ΔΔCt. Primer sequences
were 5'-TGGGATTTACCCTGTGAAACAG-3' (forward) and
5'-GACATACATGCAAAGTTCCTT-3' (reverse) for circEHBP1,
5'-AAACGGCTACCACATCCAAG-3' (forward) and 5'-TCGCGGAAGGATTTAAAGTG-3'
(reverse) for 18S rRNA, 5'-GGAUCTTTTGCGGTCTGG-3' (forward) and
5'-AGATACTTTTTGGGGTAAGGGC-3' (reverse) for premature miR-129, and
5'-CTTTTTGCGGTCTGGGCTTG-3' (forward) for mature miR-129. Universal
reverse primer and U6 primers were included in the kit.
RNA-RNA pull-down assay. In vitro
transcripts of both circEHBP1 and negative control (NC), RNA were
prepared through reverse transcriptions using HiScribe™ T7 High Yield
RNA Synthesis Kit (NEB). To perform RNA-RNA pull-down, we labeled the
3' end of both transcripts using Pierce Biotin 3' End DNA Labeling Kit
(Thermo Scientific) and re-named Bio-circEHBP1 and Bio-NC,
respectively. Bio-circEHBP1 and Bio-NC were transfected both into HL60
and HL60/ADR cells. At 48h post-transfection, cells were lysed on ice
for 10 min and incubated with magnetic beads to pull-down RNA complex.
After that, the RNA complex was purified and subject to RT-PCR to
determine the expression of circEHBP1.
Subcellular fractionation assay.
The nuclear and cytoplasm fractions of both HL60 and HL60/ADR cells
were prepared using a cell fractionation kit (ab109719, Abcam). In
brief, cells were lysed on ice for 10 min and centrifuged for 10 min at
600g. The supernatants were collected as the cytoplasm fraction. The
pellets (nuclear fraction) were further incubated with cell lysis
buffer for 10 min on ice. After that, RNAs were isolated from both
cellular fractions and subjected to RT-qPCR to determine the expression
of miR-129.
Flow cytometry analysis.
HL60 and HL60/ADR cells were harvested after the conformation of cell
transfections and incubated in a medium containing 5 μM ADR for 48h.
After that, cells were stained with Annexin V- FITC and PI and
subjected to flow cytometer S3™ Cell Sorter (Bio-Rad) to analyze cell
apoptosis.
Statistical analysis.
Two groups were compared by unpaired t-test. Two-time points of the
same group were compared by paired t-test. Multiple independent groups
were compared by ANOVA Tukey's test. The 60 AML patients were grouped
into high and low circEHBP1/mature miR-129 level groups (cutoff =
median). Chi-squared test was applied to study the associations between
circEHBP1/mature miR-129 expression and patients' clinical factors.
P<0.05 was statistically significant.
Results
Expression of circEHBP1 and miR-129 in AML patients.
The 60 patients were grouped into ADR-sensitive (n=40) and resistant
(n=20) groups based on treatment outcomes. BMMCs from ADR-sensitive
(n=40), ADR-resistant (n=20), and control (n=48) groups were subjected
to RT-qPCR to determine circEHBP1, premature miR-129 and mature miR-129
levels. Compared to the controls, AML patients exhibited increased
expression of circEHBP1 (Figure 1A, p<0.01) and premature miR-129 (Figure 1B, p<0.01) but decreased mature miR-129 (Figure 1C,
p<0.01). Altered gene expression was more obvious in the
ADR-resistant group than in the ADR-sensitive group. Association
analysis showed that circEHBP1 and mature miR-129 expression was
closely associated with patients' stages but not other factors (Table 1).
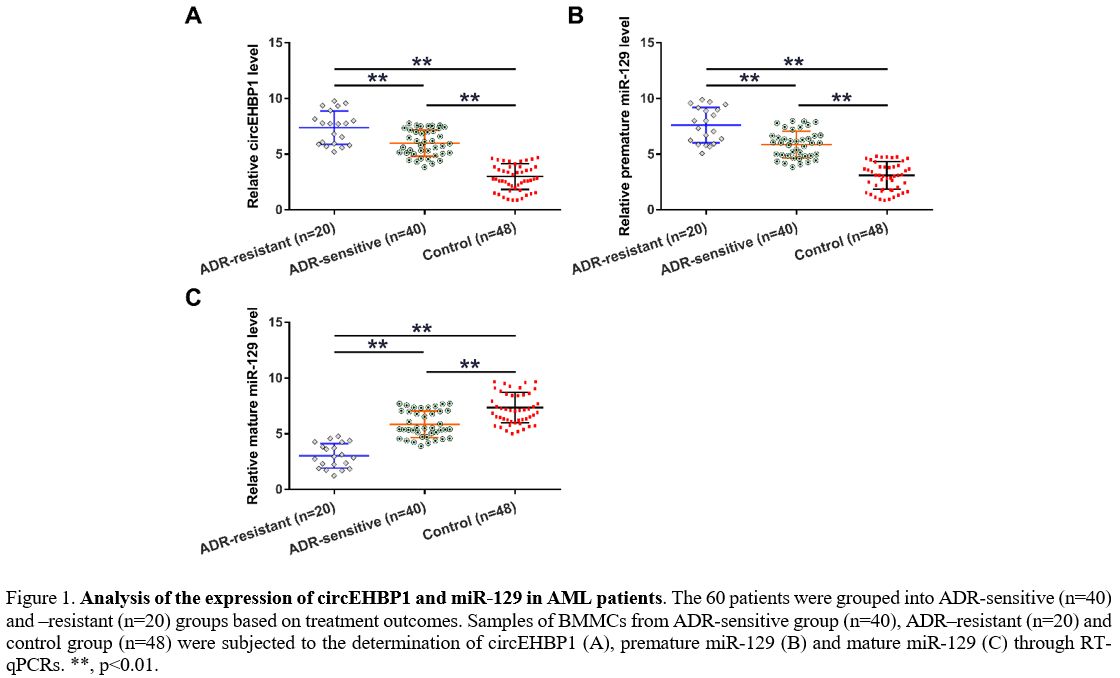 |
Figure 1. Analysis of the
expression of circEHBP1 and miR-129 in AML patients. The 60 patients
were grouped into ADR-sensitive (n=40) and –resistant (n=20) groups
based on treatment outcomes. Samples of BMMCs from ADR-sensitive group
(n=40), ADR–resistant (n=20) and control group (n=48) were subjected to
the determination of circEHBP1 (A), premature miR-129 (B) and mature
miR-129 (C) through RT-qPCRs. **, p<0.01. |
 |
Table 1. Associations between circEHBP1 and miR-129 expression and AMP patients’ clinical factors |
Expression of circEHBP1 and miR-129 in ADR-sensitive and resistant HL60 cells. IC50
of ADR was determined by MTT assay to evaluate ADR resistance of both
HL60 and HL60/ADR cells. Compared with parental HL60 cells, HL60/ADR
cells presented a poorer response to ADR, as evidenced by increased IC50 (Figure 2A,
p<0.01). ADR-sensitive and resistant HL60 cells were also subjected
to RT-qPCR to determine the expression of circEHBP1, premature miR-129,
and mature miR-129. Compared to ADR-sensitive HL60 cells, ADR-resistant
HL60 cells exhibited increased expression of circEHBP1 (Figure 2B, p<0.01) and premature miR-129 (Figure 2C, p<0.01), but decreased mature miR-129 (Figure 2D, p<0.01).
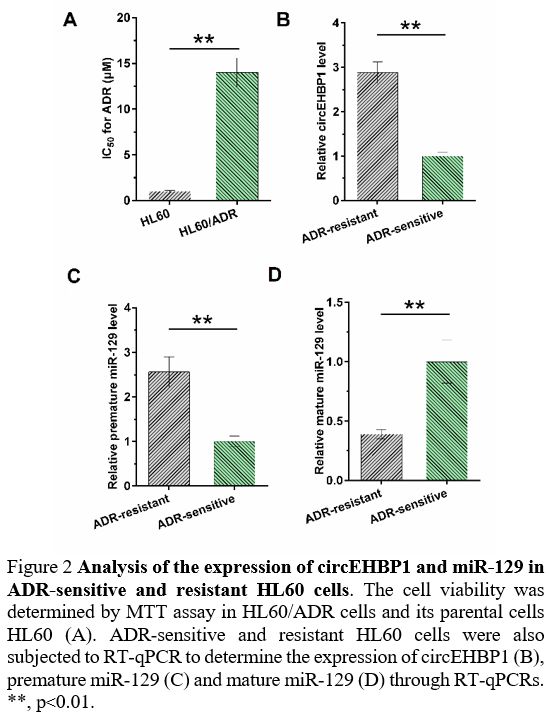 |
Figure 2. Analysis of the expression of circEHBP1 and miR-129 in ADR-sensitive and resistant HL60 cells.
The cell viability was determined by MTT assay in HL60/ADR cells and
its parental cells HL60 (A). ADR-sensitive and resistant HL60 cells
were also subjected to RT-qPCR to determine the expression of circEHBP1
(B), premature miR-129 (C) and mature miR-129 (D) through RT-qPCRs. **,
p<0.01.
|
Detection of circEHBP1 in nucleus and cytoplasm and its direct interaction with premature miR-129.
A cellular fractionation assay was carried out to detect circEHBP1 in
the nucleus and cytoplasm in both HL60 and HL60/ADR cells. It was
observed that circEHBP1 could be found in both the nucleus and
cytoplasm (Figure 3A). IntaRNA
2.0 was applied to predict the direct interaction between circEHBP1 and
premature miR-129. The prediction revealed a strong base pairing
between them (Figure 3B).
RNA-RNA pull-down assay was applied to confirm their direct interaction
further. Compared to the Bio-NC group, significantly higher levels of
miR-129 were observed in Bio-circEHBP1, establishing the direct
interaction between them (Figure 3C, p<0.01).
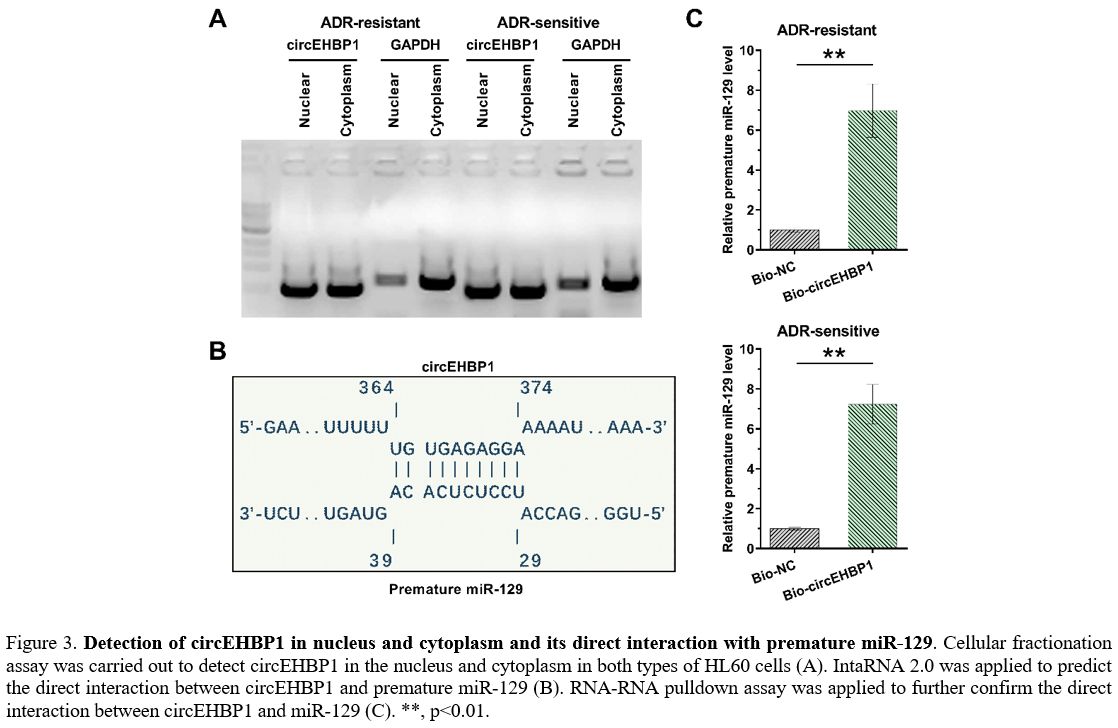 |
Figure
3. Detection of circEHBP1 in nucleus and cytoplasm and its direct interaction with premature miR-129.
Cellular fractionation assay was carried out to detect circEHBP1 in the
nucleus and cytoplasm in both types of HL60 cells (A). IntaRNA 2.0 was
applied to predict the direct interaction between circEHBP1 and
premature miR-129 (B). RNA-RNA pulldown assay was applied to further
confirm the direct interaction between circEHBP1 and miR-129 (C). **,
p<0.01. |
Analysis of the role of circEHBP1 in the maturation of miR-129.
HL60 and HL60/ADR cells were overexpressed with circEHBP1 or miR-129,
and RT-qPCR confirmed their overexpression. Furthermore, it was
observed that circEHBP1 and miR-129 were significantly overexpressed;
the overexpression of circEHBP1 was more obvious in ADR-sensitive cells
than in ADR-resistant cells, while overexpression of miR-129 was less
obvious in ADR-sensitive cells than in ADR-resistant cells (Figure 4A). In both HL60 and HL60/ADR cells, circEHBP1 increased the level of premature miR-129 (Figure 4B, p<0.01), but decreased the level of mature miR-129 (Figure 4C, p<0.01).
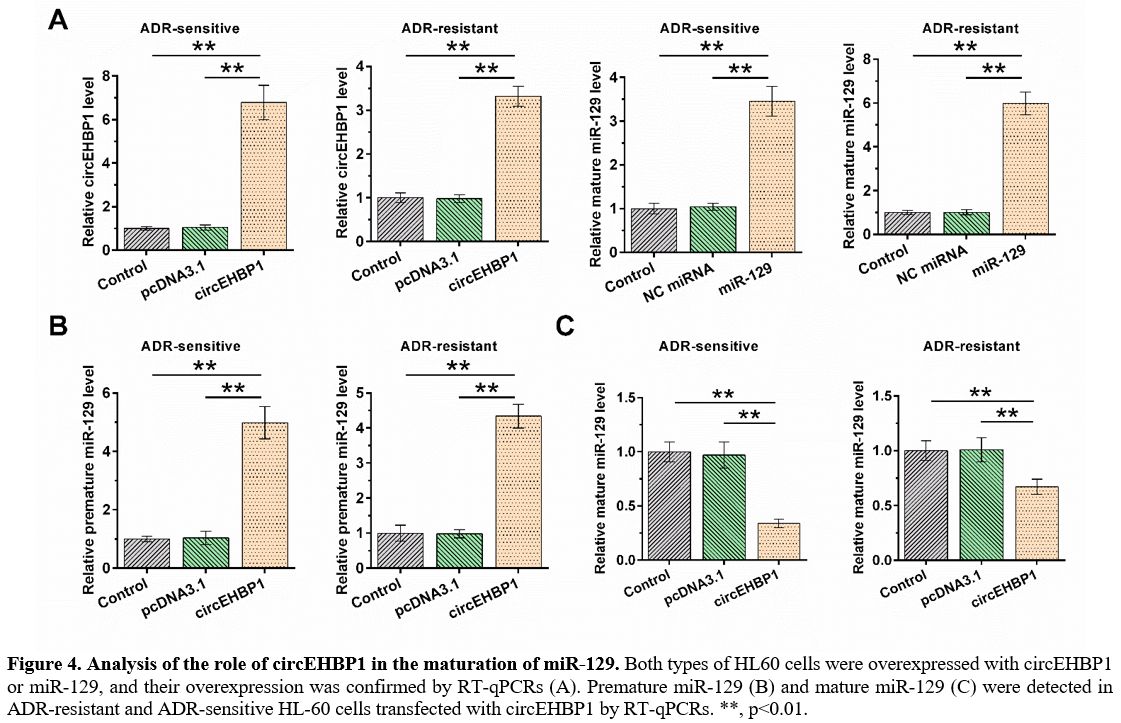 |
Figure 4. Analysis of the role of circEHBP1 in the maturation of miR-129.
Both types of HL60 cells were overexpressed with circEHBP1 or miR-129,
and their overexpression was confirmed by RT-qPCRs (A). Premature
miR-129 (B) and mature miR-129 (C) were detected in ADR-resistant and
ADR-sensitive HL-60 cells transfected with circEHBP1 by RT-qPCRs. **,
p<0.01. |
The role of circEHBP1 and miR-129 in ADR-induced cell apoptosis.
Cells with transfections were subjected to the analysis of ADR-induced
cell apoptosis. In each transfection group, cell apoptotic rates were
higher in ADR-sensitive cells than in ADR-resistance cells. Moreover,
circEHBP1 suppressed ADR-induced cell apoptosis and attenuated the
enhancing effects of miR-129 on cell apoptosis. More importantly, the
impact of circEHBP1 and miR-129 on regulating cell apoptosis was more
evident in ADR-resistance cells (Figure 5, p<0.01).
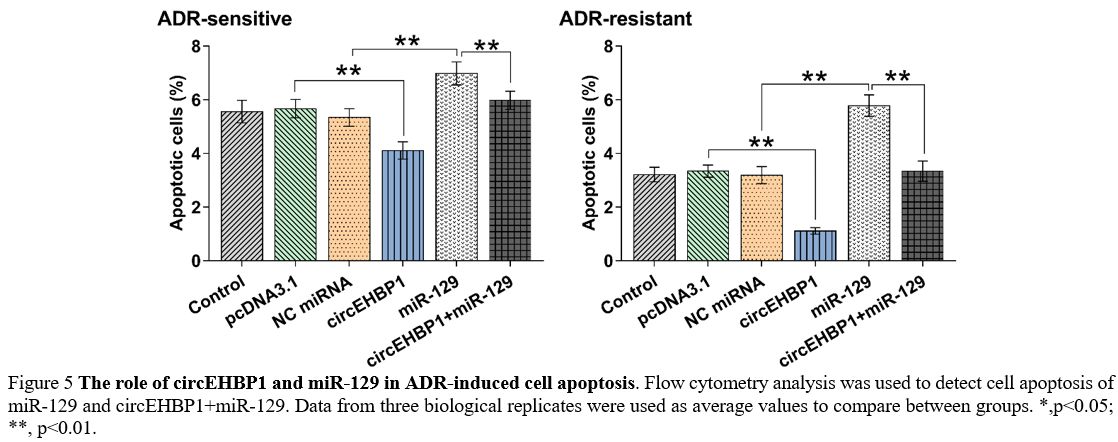 |
Figure 5. The role of circEHBP1 and miR-129 in ADR-induced cell apoptosis.
Flow cytometry analysis was used to detect cell apoptosis of miR-129
and circEHBP1+miR-129. Data from three biological replicates were used
as average values to compare between groups. *,p<0.05; **, p<0.01. |
Discussion
The
present study analyzed the expression pattern and functionality of
miR-129 and circEHBP1 in AML. We showed that circEHBP1 expression was
increased in AML patients, and the maturation of miR-129 was suppressed
in AML. Interestingly, the maturation of miR-129 was found to be
regulated by circEHBP1.
Previous studies have characterized
miR-129 as a critical player in the development of chemoresistance of
cancer cells to multiple chemical drugs.[16-18] For
instance, miR-129 was downregulated in colorectal cancer, and its
overexpression increased the sensitivity of cancer cells to
5-fluorouracil.[16] In addition, miR-129 suppressed
neuroblastoma growth, and overexpression of miR-129 increased the
sensitivity of cancer cells to Cytoxan via MYO10.[17] In gastric cancer, miR-129 targeted P-gp to suppress cisplatin resistance.[18]
Based on our knowledge, the role of miR-129 in AML is unclear. In this
study, we showed the upregulation of premature miR-129 and
downregulation of mature miR-129, and the alteration was more obvious
in ADR-resistant patients than in ADR-sensitive patients. Therefore,
inhibition of miR-129 maturation is likely involved in AML. Moreover,
overexpression of miR-129 increased the sensitivity of AML cells to
ADR. Thus, miR-129 may be targeted to raise the chemosensitivity of AML
cells to ADR.
A recent study reported that circEHBP1 promoted
bladder cancer progression by increasing lymphatic metastasis and
lymphangiogenesis through its interaction with
miR-130a-3p/TGFβR1/VEGF-D.[19] The present study
characterized the expression pattern of circEHBP1 and observed its
upregulation in AML. Moreover, circEHBP1 overexpression suppressed
ADR-induced cell apoptosis. Therefore, circEHBP1 is likely an oncogenic
circRNA in AML. Most importantly, we detected circEHBP1 in both nuclear
and cytoplasm fractions of AML cells, and circEHBP1 directly interacted
with premature miR-129.
Moreover, circEHBP1 overexpression
suppressed miR-129 maturation. Therefore, premature miR-129 may be
sponged by circEHBP1 in the nucleus, reducing its maturation. It is
worth noting that circEHBP1 was predicted to interact with multiple
premature miRNAs (data not shown), while our functional assays only
validated its interaction with premature miR-129. Interestingly,
although the effects of circEHBP1 and miR-129 are stronger in
ADR-resistant cells than in ADR-sensitive cells, they regulated
ADR-induced apoptosis in both cell types. Therefore, circEHBP1 and
miR-129 can control the sensitivity of cancer cells in both
ADR-resistant and sensitive patients.
Conclusions
CircEHBP1
was overexpressed in AML, and the maturation of miR-129 was suppressed
in AML. In addition, circEHBP1 may suppress miR-129 maturation to
decrease ADR-induced cell apoptosis.
References
- Short NJ, Rytting ME, Cortes JE. Acute myeloid leukaemia. Lancet (London, England). 2018;392(10147):593-606. https://doi.org/10.1016/S0140-6736(18)31041-9
- Abelson
S, Collord G, Ng SWK, et al. Prediction of acute myeloid leukaemia risk
in healthy individuals. Nature. 2018;559(7714): 400-404. https://doi.org/10.1038/s41586-018-0317-6 PMid:29988082 PMCid:PMC6485381
- Khwaja A, Bjorkholm M, Gale RE, et al. Acute myeloid leukaemia. Nature Reviews Disease Primers. 2016;2:16010. https://doi.org/10.1038/nrdp.2016.10 PMid:27159408
- Ferrara F, Schiffer CA. Acute myeloid leukaemia in adults. Lancet (London, England). 2013;381(9865):484-495. https://doi.org/10.1016/S0140-6736(12)61727-9
- Shah
A, Andersson TM, Rachet B, Björkholm M, Lambert PC. Survival and cure
of acute myeloid leukaemia in England, 1971-2006: a population-based
study. British Journal of Haematology. 2013;162(4):509-516. https://doi.org/10.1111/bjh.12425 PMid:23786647
- Krug
U, Röllig C, Koschmieder A, et al. Complete remission and early death
after intensive chemotherapy in patients aged 60 years or older with
acute myeloid leukaemia: a web-based application for prediction of
outcomes. Lancet (London, England). 2010;376(9757):2000-2008. https://doi.org/10.1016/S0140-6736(10)62105-8
- Kayser
S, Levis MJ. Advances in targeted therapy for acute myeloid leukaemia.
British Journal of Haematology. 2018;180(4):484-500. https://doi.org/10.1111/bjh.15032 PMid:29193012 PMCid:PMC5801209
- Barrett
AJ, Le Blanc K. Immunotherapy prospects for acute myeloid leukaemia.
Clinical and Experimental Immunology. 2010;161(2):223-232. https://doi.org/10.1111/j.1365-2249.2010.04197.x PMid:20529084 PMCid:PMC2909404
- Yi
Z, Gao K, Li R, Fu Y. Dysregulated circRNAs in plasma from active
tuberculosis patients. Journal of Cellular and Molecular Medicine.
2018;22(9):4076-4084. https://doi.org/10.1111/jcmm.13684 PMid:29961269 PMCid:PMC6111852
- Sun
YM, Wang WT, Zeng ZC, et al. circMYBL2, a circRNA from MYBL2, regulates
FLT3 translation by recruiting PTBP1 to promote FLT3-ITD AML
progression. Blood. 2019;134(18):1533-1546. https://doi.org/10.1182/blood.2019000802 PMid:31387917 PMCid:PMC6839953
- Han
F, Zhong C, Li W, et al. hsa_circ_0001947 suppresses acute myeloid
leukemia progression via targeting hsa-miR-329-5p/CREBRF axis.
Epigenomics. 2020;12(11):935-953. https://doi.org/10.2217/epi-2019-0352 PMid:32657138
- Hu
Q, Gu Y, Chen S, Tian Y, Yang S. Hsa_circ_0079480 promotes tumor
progression in acute myeloid leukemia via miR-654-3p/HDGF axis. Aging.
2020;13(1):1120-1131. https://doi.org/10.18632/aging.202240 PMid:33290265 PMCid:PMC7835062
- Zhu
J, Luo Y, Zhao Y, et al. circEHBP1 promotes lymphangiogenesis and
lymphatic metastasis of bladder cancer via miR-130a-3p/TGFbetaR1/VEGF-D
signaling. Molecular therapy : the Journal of the American Society of
Gene Therapy. 2021;29(5):1838-1852. https://doi.org/10.1016/j.ymthe.2021.01.031 PMid:33545359 PMCid:PMC8116613
- Kinoshita
C, Aoyama K. The Role of Non-Coding RNAs in the Neuroprotective Effects
of Glutathione. International Journal of Molecular Sciences.
2021;22(8). https://doi.org/10.3390/ijms22084245 PMid:33921907 PMCid:PMC8073493
- Liao Q, Wang B, Li X, Jiang G. miRNAs in acute myeloid leukemia. Oncotarget. 2017;8(2):3666-3682. https://doi.org/10.18632/oncotarget.12343 PMid:27705921 PMCid:PMC5356910
- Karaayvaz
M, Zhai H, Ju J. miR-129 promotes apoptosis and enhances
chemosensitivity to 5-fluorouracil in colorectal cancer. Cell Death
& Disease. 2013;4(6):e659. https://doi.org/10.1038/cddis.2013.193 PMid:23744359 PMCid:PMC3702282
- Wang
X, Li J, Xu X, Zheng J, Li Q. miR-129 inhibits tumor growth and
potentiates chemosensitivity of neuroblastoma by targeting MYO10.
Biomedicine & Pharmacotherapy = Biomedecine & Pharmacotherapie.
2018;103:1312-1318. https://doi.org/10.1016/j.biopha.2018.04.153 PMid:29864913
- Lu
C, Shan Z, Li C, Yang L. MiR-129 regulates cisplatin-resistance in
human gastric cancer cells by targeting P-gp. Biomedicine &
Pharmacotherapy = Biomedecine & Pharmacotherapie. 2017;86:450-456. https://doi.org/10.1016/j.biopha.2016.11.139 PMid:28012924
- Zhu
J, Luo Y, Zhao Y, et al. circEHBP1 promotes lymphangiogenesis and
lymphatic metastasis of bladder cancer via miR-130a-3p/TGFβR1/VEGF-D
signaling. Molecular Therapy: the Journal of the American Society of
Gene Therapy. 2021;29(5):1838-1852. https://doi.org/10.1016/j.ymthe.2021.01.031 PMid:33545359 PMCid:PMC8116613,
[TOP]





