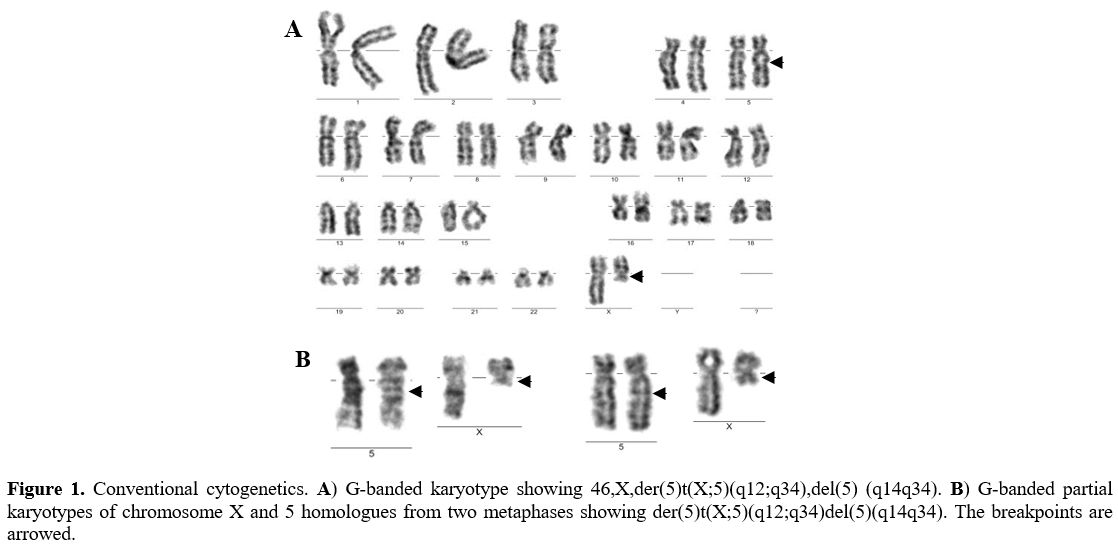
A 73-year-old woman was referred to the clinic with complaints of easy fatigue and a history of pneumonia from the previous year. Laboratory tests showed hemoglobin 90g/l, mean corpuscular volume 98fl, erythrocyte sedimentation rate (ESR) 23мм, white blood cells (WBC) 4.5x109/l (neutrophils 3.51x109/l) and platelets 87x109/l. The X-ray found fibrocalcific changes in both hilus of the lung. The bone marrow examination showed normocellular marrow with 1% blasts, megaloblastic erythropoiesis, hypo- or agranular mature neutrophils (22%), and an increased number of megakaryocytes (1%) with prominent dysplasia (38% hypo- or non-lobated micromegakaryocytes). Conventional cytogenetics revealed the following new clonal chromosome rearrangement: 46,X,t(X;5)(Xpter=>Xq12::5q34=>5qter;5pter>5q14::Xq12=>Xqter)[20]/46,XX[5] (Figure 1).
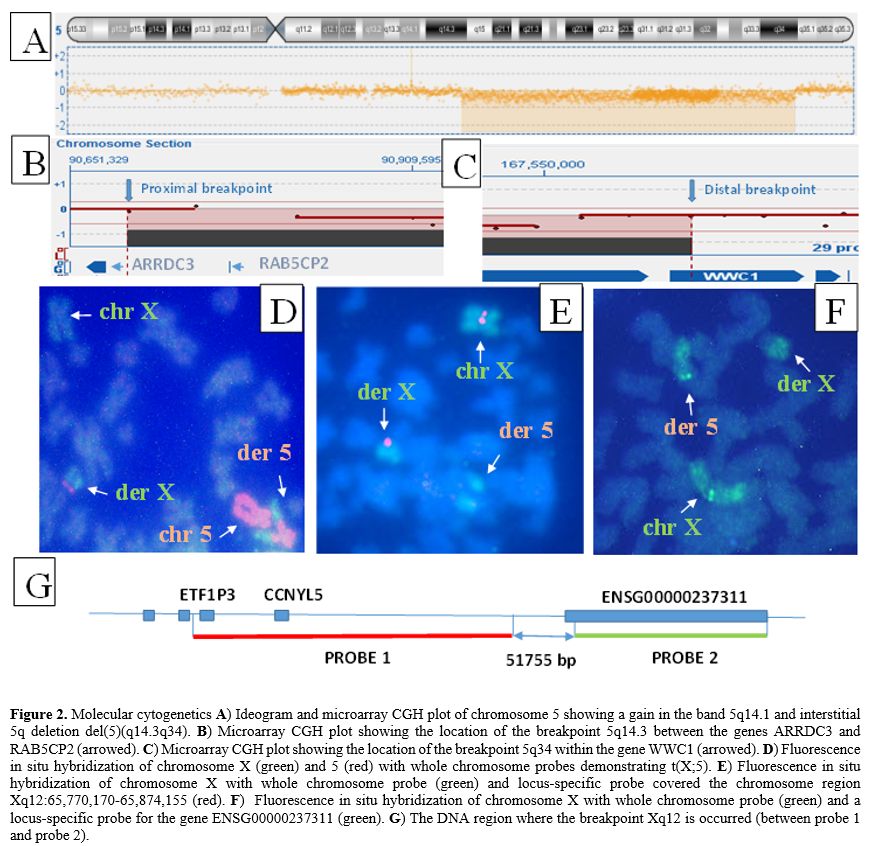
No stem line with del(5)(q14q34) was detected, which suggests that 5q- and t(X;5) have arisen simultaneously. Fluorescence in situ hybridization (FISH) with whole chromosome probes for X and 5 confirmed t(X;5):46, X.ish t(X;5)(q12;q34)(wcpX+,wcp5+;wcp5+.wcpX+) (Figure 2D). Microarray comparative genomic hybridization with 60K microarray platform (OGT, Oxford, UK) established two unbalanced anomalies in 5q: gain at band 5q14.1 and loss at 5q14.3 through 5q34 which are with 367.91Kb and 77.05Mb in size: arr[hg19] 5q14.1(79,161,513-79,529,419)x3,5q14.3q34(90,695,096-167,742,618)x1 (Figure 2A). The breakpoint in 5q14.3 occurred in the noncoding region between the genes ARRDC3 and RAB5CP2 (Figure 2B) and in 5q34 within the WWC1 gene (Figure 2C) (validated by FISH). To determine the breakpoint location on Xq12 FISH experiments were performed with whole staining of the X chromosome combined with two locus-specific probes - one covered the chromosome region Xq12:65,770,170-65,874,155 (104kb; hg38) and another the gene ENSG00000237311 (Xq12:65,925,930-66,001,435;hg38) (Agilent Technologies, Santa Clara, CA, USA). The signal from the first probe was found on the X chromosome (Figure 2E), while the signal from the second probe was on chromosome 5 (Figure 2F). These data revealed that the breakpoint on Xq12 occurred in the 52kb region (56,925,930-66,001,435) (hg38) that included 47.5kb noncoding DNA sequences and 4.5 kb coding sequences from the 5' end of the ENSG00000237311 gene (Figure 2G). Collectively, the genomic studies showed that 1) del(5)(q14.3q34) resulted in a loss of the whole RAB5CP2 gene and part of the WWC1 gene (the size of the lost part is 23862bp and included the enhancer, the WW domains, and the first and second promoter) and 2) t(X;5) caused juxtaposition of the deleted WWC1 gene to the X chromosome, and ENSG00000237311 gene to chromosome 5 respectively (near to ARRDC3 gene) (Figure 3). The diagnosis of 5q- syndrome was made, and therapy with lenalidomide was started at a reduced dose of 5 mg daily because of thrombocytopenia. Over the next two months, along with pancytopenia caused by the lenalidomide, a gradual increase of monocytes, ESR, and C-reactive protein (CRP) was observed. At the end of the third month, the patient was re-admitted to the clinic with a deteriorating general condition, low-grade fever (37.2Co), pancytopenia (hemoglobin 70g/l, WBC 2.1x109/l, and platelets 52x109/l), neutropenia (0.39x109/l), relative monocytosis (33%, 0.7x109/l) and increased inflammation markers (ESR 70 mm and CRP 17.8 mg/l). Body examination found two enlarged cervical lymph nodes (2.5 cm in diameter) in the supraclavicular area, which were movable, painless, and firm in consistency. Histopathology after excisional lymph node biopsy revealed granulomatous lymphadenitis with epitheloid and giant cells. QuantiFERON-TB gold test was positive and confirmed the diagnosis of extrapulmonary TB manifestеd as tuberculosis lymphadenitis. The clinical condition was considered a reactivation of latent TB into active TB caused by lenalidomide treatment.[2] The therapy with lenalidomide was discontinued, and an anti-TB medication with ethambutol 15 mg/kg, isoniazid 300 mg/day, rifampicin 600 mg/day, and pyrazinamide 15 mg/kg was started. After six months of anti - TB therapy, the patient's lymphadenopathy disappeared, and the number of neutrophils and platelets increased to 1,7x109/l and 80x109/l, respectively. Monocytes, ESR, and CRP, decreased to normal values. Lenalidomide treatment was re-initiated at a reduced dose of 5 mg every other day. During the next four years, the patient maintained transfusion independence (hemoglobin up to 100 g/l) and good quality of life.
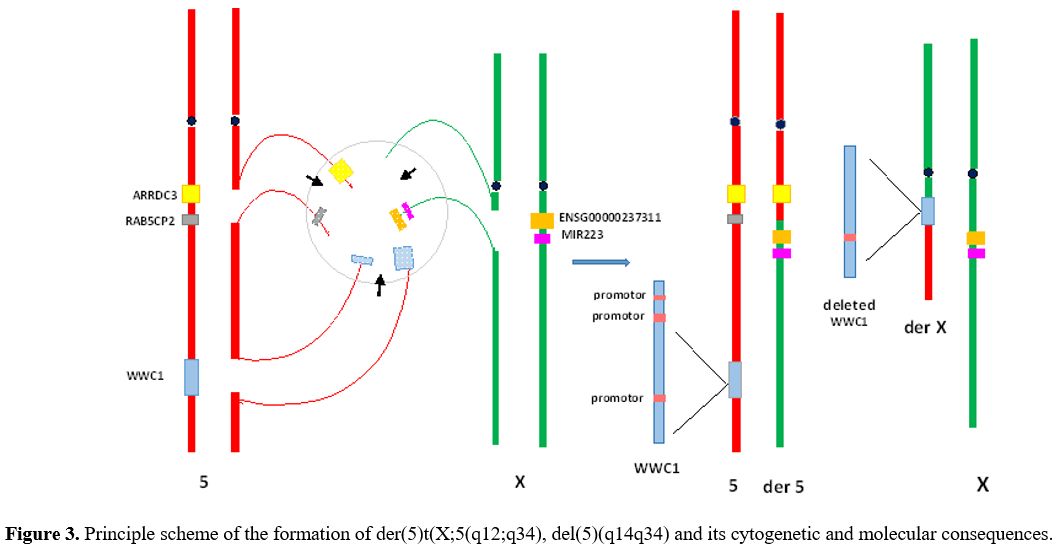 |
Figure 3. Principle scheme of the formation of der(5)t(X;5(q12;q34), del(5)(q14q34) and its cytogenetic and molecular consequences. |
The functions of the described genes will clarify the cellular events that have provoked der(5)t(X;5),del(5q). ARRDC3 is a member of the alpha arrestins that mediate endosome trafficking leading to recycling or lysosomal degradation of G protein-coupled receptors.[3] The second gene, RAB5CP2, is a pseudogene. Its possible function as a pseudogene is to regulate the expression of its parent gene[4] RAB5C, which is a regulator of the early stage of the endocytic pathway.[5] The protein of the third gene WWC1 (Kibra) (5q34) as part of the protein complexes dynein/SNX4 and exocyst/PKCzeta mediate the direction of transferrin receptor to a recycling pathway and transcriptional co-activation of estrogen receptor – alpha[6,7] respectively. Kibra is also involved in the regulation of autophagosomes, promoting autophagosomal degradation via the Hippo pathway (through its WW domains).[8] The data from these three genes suggest an obvious regularity - the proteins encoded by them are related to the endocytic and phagosomal pathways that are closely interconnected and are involved in cellular homeostasis, including the protection of organisms from invading pathogens. It is also interesting that the located at Xq12 gene ENSG00000237311 is a long noncoding RNA with unknown function, but close to it is mapped miR-223, whose increased expression has been found in both 5q- syndrome and tuberculosis.[9,10] Moreover, miR-223 is critical in mediating the innate host response to tuberculosis.[11] Finally, it is logical to suppose that, in our case, the loss of RAB5CP2 from the genome and the deletion of WW domains of WWC1 can affect the maturation of the early Rab 5 phagosomes and the hypo pathway with subsequent blocking of the lysosomal degradation of the pathogen. The consequences of the described anomaly could favor the cellular adaptation of the bacterial pathogen, consistent with the reports demonstrating that Mycobacterium tuberculosis can manipulate early Rab 5 phagosomes and the Hippo pathway to modulate the host immune response.[10,12]
In conclusion, the appearance of the established clonal abnormality in our case is probably linked to the signaling network induced by the bacterial pathogen to avoid the host's innate immune system, which possibly includes also sex-dependent mechanisms such as deregulation of noncoding RNA genes located on the X chromosome.