 |
|
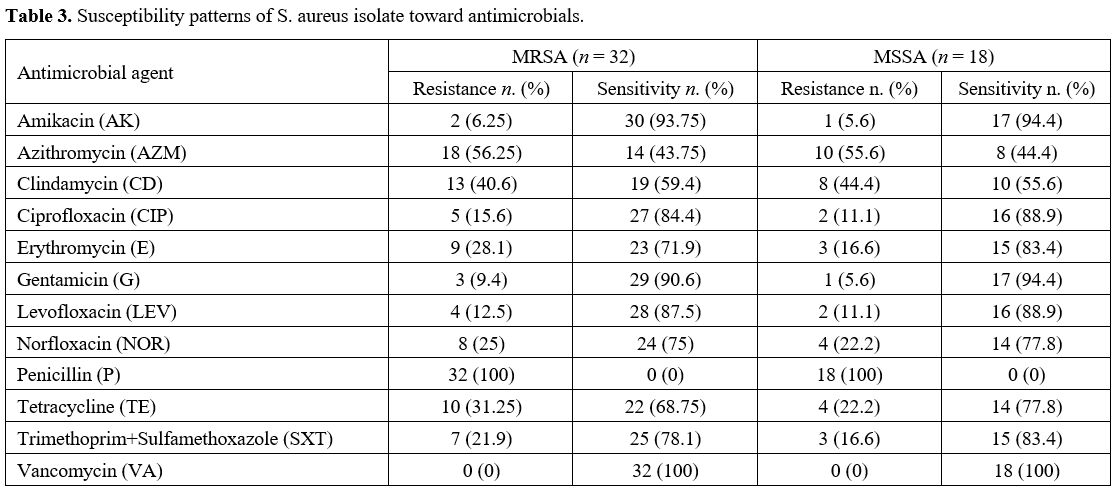
Antibiotic Susceptibility Testing. The disc diffusion technique was used to test all staphylococcal isolates for methicillin resistance. PCR verified methicillin resistance to detect the mecA gene.[16] The isolates' susceptibility was evaluated using the disc diffusion technique[20] using the following antibiotic discs (Bioanalyse, Turkey): Amikacin AK 30 μg, Azithromycin AZM 15 μg, Clindamycin CD 2 μg, Ciprofloxacin CIP 5 μg, Erythromycin E 15 μg, Gentamicin G 10 μg, Levofloxacin LEV 5 μg, Norfloxacin NOR 10 μg, Penicillin P 10 U, Tetracycline TE 30 μg, Trimethoprim+Sulfamethoxazole SXT 1.25+23.75 μg, and Vancomycin VA 30 μg. S. aureus ATCC 25923 was used as the control strain.
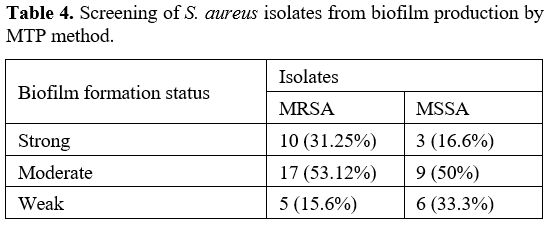
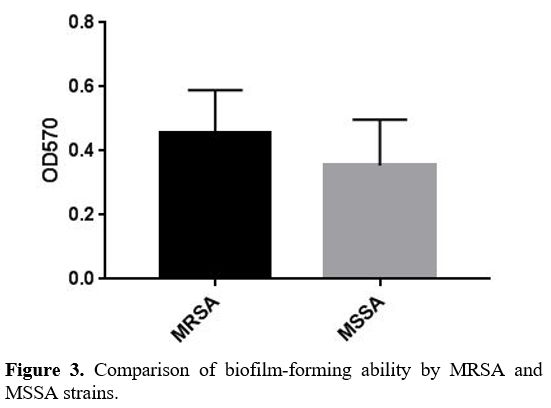
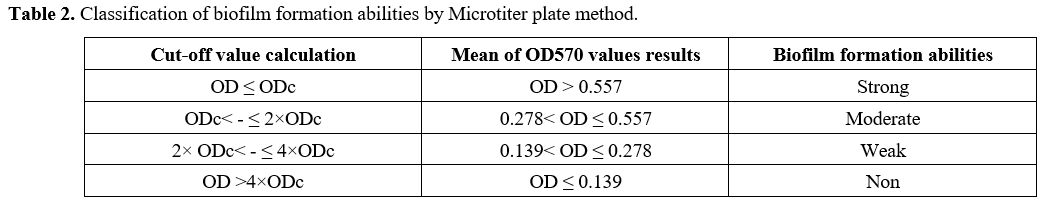
Biofilm Formation Assay. Biofilm generation was quantified using a Microtiter plate (MTP) approach described by Yousefi M. et al.[21] In brief, bacterial isolates were cultured in trypticase soy broth (TSB) (Merck, Germany) with 0.5 percent glucose and incubated at 37ºC overnight. Cultures with 0.5 percent glucose with 1:40 in fresh TSB were diluted (Sigma, USA). Two hundred μL of the diluted solution was put into Microtiter plate wells and incubated for 48 hours at 37ºC. Only 200 μL of TSB-0.5% glucose was present in the negative control wells, and there was no bacterial suspension. Wells were carefully cleaned three times with phosphate buffer saline (PBS) (pH 7.2), fixed for 20 minutes with methanol, dried at room temperature, and stained for 10 minutes with crystal violet 0.1 percent. Then 1 mL of 95% ethanol was added to each well to dissolve the dye bound to the adherent cells. Finally, using an ELISA reader (BioTek ELx800, USA), optical density (OD) at 570 nm (A570) was determined for each well. The average OD of negative control + 3 standard deviation (SD) of negative control was calculated for the optical density cut–off (ODc). Based on the absorbance of crystal violet stain linked to the adhered cells, biofilms formed by various strains have been analyzed and categorized (Table 2).
 |
|
DNA Isolation. DNA extraction from a genome following the manufacturer's instructions, genomic DNA was isolated from pure cultures using the Presto™ Mini gDNA Bacteria Kit (Geneaid, Taiwan); the extract was then eluted with a 100 μL elution buffer. Extracts were kept at -20°C until PCR was performed. A DNA template was used for PCR using 2 μL of the total extracted material from each test sample.
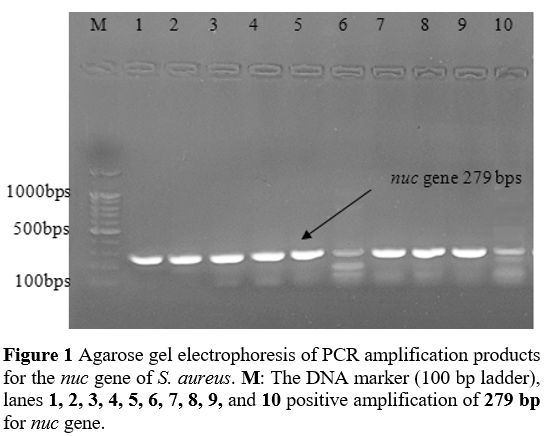
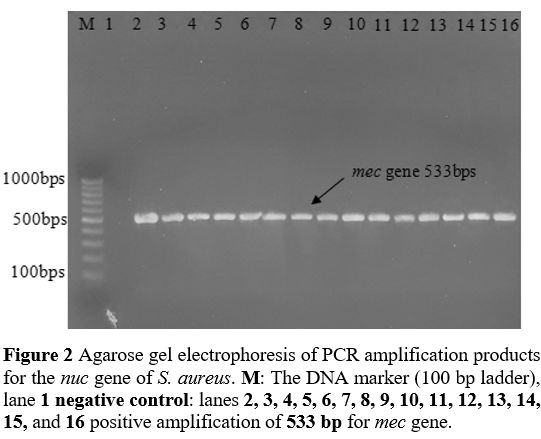
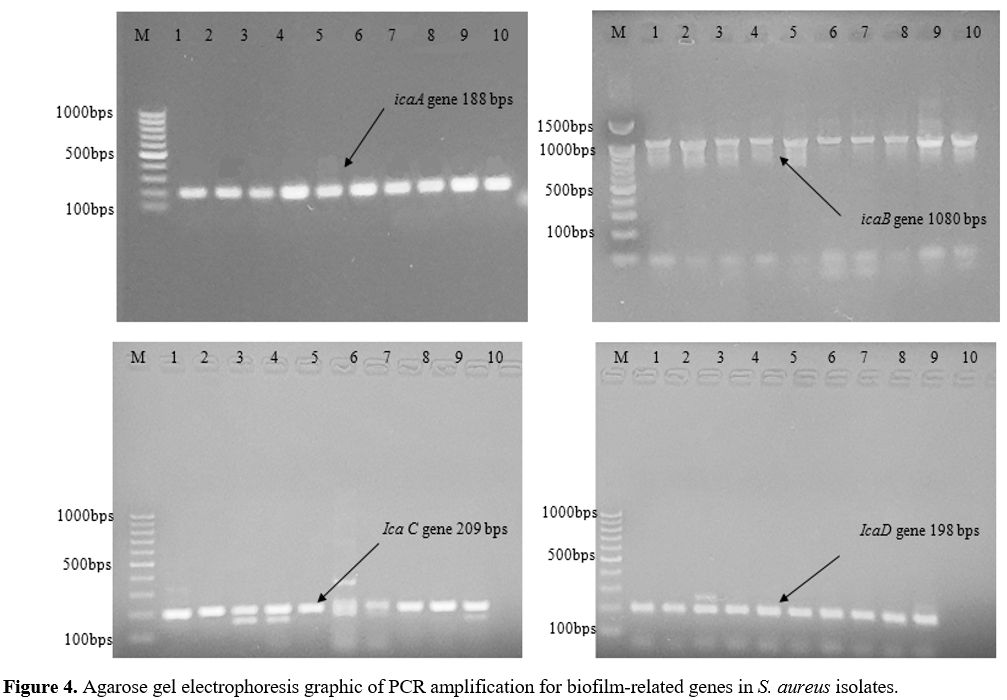
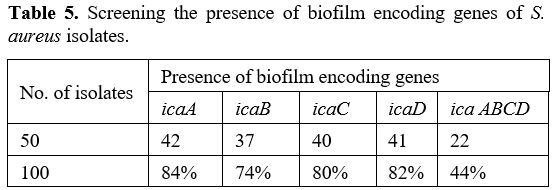
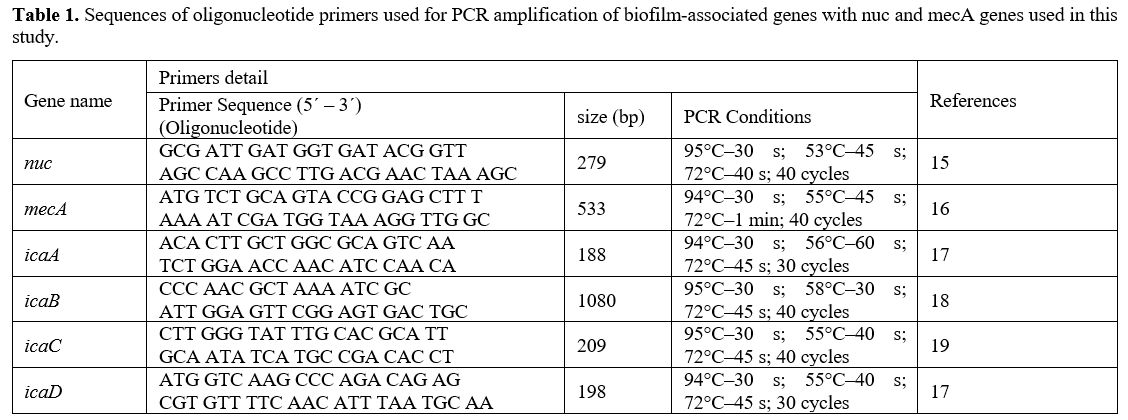
Primers and PCR Conditions. The primers designed by BioGene (South Korea) specifically for the nuc, mecA, icaA, icaB, icaC, and icaD are given in Table 2. The monoplex PCR was performed in a 25-𝜇L volume for each gene. All PCR reactions were conducted by employing 2 μl DNA template (density of 10 ng/μl), a Master Mix comprising of 3 mM MgCl2, 0.2% Tween® 20, 20 mM Tris-HCl pH 8.5, (NH4)2S04, 0.4 mM of each dNTP, 0.4 μM of each primer, and 0.2 units/μl Ampliqon Taq DNA polymerase. The PCR amplification conditions were established as shown in Table 1.
Statistical Analysis. The T-test was used to examine group differences at the 0.05 level of significance. Utilizing the statistical analysis program GraphPad Prism 7.