Original Articles
Prognostic
Significance of NRAS Gene Mutations in Children with Acute Myelogenous
Leukemia.R.M. Aly1, MD, M.R. El-sharnoby2 and A.A. Hagag2
1Clinical
Pathology Department, Faculty of Medicine, Mansoura University, Egypt. 2Pediatrics
Department, Faculty of Medicine, Tanta
University, Egypt.
Correspondence
to: Rabab M. Aly, MD.
Lecturer of Clinical Pathology, Faculty of Medicine,
Mansoura University, Egypt. e-mail: rababzeadah@yahoo.com
Published: November 28, 2011
Received: July 16, 2011
Accepted: October 21, 2011
Mediterr J Hematol Infect Dis 2011, 3(1): e2011059, DOI 10.4084/MJHID.2011.055
This article is available from: http://www.mjhid.org/article/view/8841
This is an Open Access article
distributed under the terms of the
Creative Commons Attribution License
(http://creativecommons.org/licenses/by/2.0), which permits
unrestricted use, distribution, and reproduction in any medium,
provided the original work is properly cited
Abstract
NRAS mutations are the most commonly detected molecular abnormalitiesin hematologic malignancies, especially in those of myeloid origin.
Objective: We aimed to determine the frequency of NRAS (NRASmutant) mutation; and its prognostic significance in Egyptian children with acute myelogenous leukemia (AML).
Subject and methods: Peripheral blood and bone marrow (BM) samples were taken from 39 de novo pediatric AML patients. Twenty subjects with matched age and sex were selected as a control group. Samples from patients and control were analyzed for Exons 1, 2 of NRAS gene using genomic PCR-SSCP method.
Results: NRAS mutations at the time of diagnosis was found in 6/39 (15.4%) AML cases. Patients with NRASmutant had no significant improved clinical outcome than patients without mutation. Patients with NRASmutant had similar complete remission (CR) rates compared with non-mutated patients (66.7% vs. 69.5%, P=0.43). Those in CR had a similar relapse rate regardless of the presence of NRASmutant (RR 33.4% vs. 30.2%, P=0.26). However, an adverse prognosis for 3 year overall survival (OS) was associated with the presence of NRAS mutations. This adverse prognosis associated with NRAS mutations was also observed in terms of disease-free survival (DFS) (P=0.007). Univariate analysis showed that unfavorable prognostic factors for DFS were cytogenetic data (P = 0.005) and the NRAS gene mutation (P = 0.002).
Conclusion: NRASmutant did not contribute to increase the disease recurrence, however NRASmutant was found to be a poor prognostic factor for children with AML. Further studies to confirm these findings are required because of the small number of patients with NRAS mutation.
Introduction
Acute myeloid leukemia (AML) is characterized by expansion of myeloid blasts with suppression of normal hematopoiesis. Cytogenetic and molecular studies have defined AML as a heterogeneous disease.[1] The presence of defined karyotypes is among the most important prognostic factors in acute myeloid leukemia (AML). However, even within defined cytogenetic groups stability of remission and long-term survival may vary significantly. Therefore, additional recurrent aberrations may have a prognostic impact.[2] It has been showed that Pediatric AML patients may harbor more than one mutation at diagnosis, some of which with a possible prognostic impact.[3-7] Mutations in the NRAS gene are one of these genetic aberrations that play a role in myeloid neoplasia.[8]
NRAS gene plays an important role in the regulatory processes that govern proliferation, differentiation and apoptosis; [9] abnormality in this gene has been implicated in the pathogenesis of AML. RAS oncogenes encode a family of membrane-associated proteins, which regulate signal transduction upon binding to a variety of membrane receptors.[10] There are three functional RAS genes (NRAS, KRAS and HRAS); in AML. K-RAS mutation occurs to a lower but still significant frequence in pediatric AML.[11] NRAS is the most prominent; reported in 11%-30% of patients.[12] All homologs were exclusively in codons [12,13], and 61 conferring constitutive activation of the RAS protein, which is subsequently held in the GTP bound status leading to an increased activity of the RAS pathway causing an increased proliferation and a decreased apoptosis rate.[13] RAS mutations were described in the various solid tumors as well as in hematologic malignancies. The prognostic impact of NRAS mutations is still under research and seems to vary from disease to disease,14 several studies indicated a poor prognostic impact for that mutation,[14,15] and however Lapillonne et al confirmed this finding in pediatric AML.[16] On the contrary, Neubauer et al found a favorable outcome for malignancies with NRAS mutations,12 and some studies failed to define any prognostic impact for NRAS mutations.[10,13,17]
We undertook this study to determine the frequency of NRAS mutation and its prognostic significance in a number of Egyptian pediatric patients with AML.
Patients and Methods
Newly diagnosed pediatric AML patients were included in this study; cases were recruited from pediatric hematology clinics of Tanta University Hospitals, Tanta, Egypt. Peripheral blood and BM samples were obtained from 39 de novo AML cases at initial diagnosis obtaining an informed consent from patients or their guardians; they were 21 boys and 18 girls. The median age was 7.4 years (range, 5.6-13 years). The median percentage of blasts in the fresh bone marrow samples was (65%). All included patients were receiving the same treatment protocol approved by the Oncology Team of Tanta University Hospital TUH; in brief they received 1-2 cycles of 14-21 days of intensively timed induction chemotherapy (doxorubicin, Ara-C, 6-thioguanine and methotrexate) depending upon BM aspiration done at the end of each induction course. Additional consolidation regimens included 1-2 cycles of 12-days (doxorubicin, Ara-C, VP-16 and methotrexate). Then patients who did not achieve remission received intermittent chemotherapy (Ara-C, 6-thioguanine and methotrexate) every 3 months for 6 cycles with the standard follow-up care and regular BM aspiration every 21 days to confirm remission; complete remission means normocellular bone marrow contain less than 5% blast cells and showing evidence of normal maturation of other bone marrow elements as evidenced by the repeated BM aspiration. The average duration of follow up was mean ± SD (32 ± 2.24 months).
Patients were classified according to the standard methods; morphological according FAB classification, cytochemical and immunological evaluation.[13] Informed consent was obtained from twenty subjects with matched age and sex who were selected as a control group. Samples from patients and control were analyzed for mutation in Exons 1, 2 of the NRAS gene using genomic PCR method.
Cytogenetic Analysis
Cytogenetic investigations were performed by karyotyping G-banding analysis in all patients.[18]
PCR of NRAS Gene
Genomic DNA was extracted from diagnostic bone marrow specimens of patients and control using the QIAamp DNA blood mini kit for DNA extraction provided by QIAGEN (Inc Chasworthy, CA). The concentration of extracted DNA was then measured by UV spectrophotometry at 260 & 280 nm and analyzed by electrophoresis on 2% agarose gel for detection of purity.
Separate assays were developed for mutation detection at (hot spots) in codons 12/13 (exon 1) and codon 61 (exon 2). Oligonucleotide primers amplifying short fragments {241 base pair (bp) for exon 1 and 201 bp for exon 2} were designed for PCR as follows:
N12/13 assay. forward, 5'-GACTGAGTACAAACTGGTGG-3'; and reverse, 5'-TGCATAACTGAATGTATACCC-3'.
N61 assay. forward, 5' CAAGTGGTTATAGATGGTGAAACC-3'; and reverse, 5'-AAGATCATCCTTTCAGAGAAAATAAT-3'.
PCR amplification was performed in 50 ul reaction, contained 50-100 ng of genomic DNA, 10 mM Tris HCl (pH 8.3), 50 mM KCl and 1.5 mM Mg Cl2, 200 uM of each deoxyribonucleotide triphosphate (dNTP), 2.5 units Taq polymerase, and 6% dimethylsulphoxide. PCR conditions were as follows: (1) Exon 1, HotStarTaq (Qiagen, Valencia, CA), and 0.625 U. Primers (12.5 pmol), N12/13 forward, N12/13 reverse. Denaturing, 95 C for 15 minutes and 94°C for 30 seconds; annealing, 55.5°C for 1minute; extension, 72°C for 1 minute for 35 cycles; final cycle at 72°C for 10 minutes. (2) Exon 2, HotStarTaq (Qiagen), 0.625 U. Primers (12.5 pmol), N61F forward, N61R reverse. Denaturing, 95°C for 15 minutes; 94°C for 30 seconds; annealing, 55.5°C for 1 minute; extension, 72°C for 1 minute for 35 cycles; final cycle at 72°C for 10 minutes.
Single-strand conformation polymorphism (SSCP) was performed to PCR product to detect mutations. Products were mixed with 10 volumes of loading buffer, quenched on ice immediately, and applied to 5% polyacrylamide gel electrophoresis at 50 V, overnight, stained by silver nitrates and wrapped in plastic foil. Normal gene exhibits a specific conformational pattern, while mutant gene displays pattern with different electrophoretic mobility (mobility shift) which was confirmed by repeated SSCP (Figure 1).
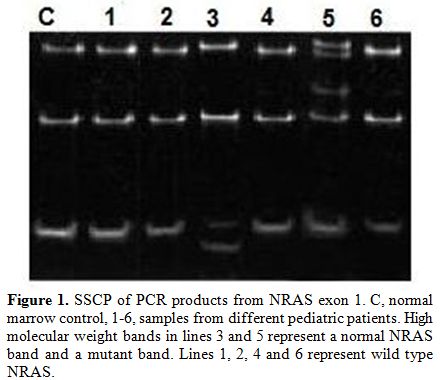
Figure 1. SSCP of PCR products from NRAS exon 1. C, normal marrow control, 1-6, samples from different pediatric patients. High molecular weight bands in lines 3 and 5 represent a normal NRAS band and a mutant band. Lines 1, 2, 4 and 6 represent wild type NRAS.
Statistical Methods
Data were processed and analyzed using SPSS for windows version 16.0 (SPSS, Inc, Chicago, IL, USA). Qualitative data were expressed as frequency and percentage and quantitative data were expressed as median. Chi-square test was used for comparative analysis. The prevalence of NRAS mutations in AML was too low to permit statistical analysis for correlation with survival. Kaplan-Meier analysis was used for survival of patients. The prognostic significance of the clinical variables was assessed using the Cox proportional hazards model. For all analyses, the P values were two-tailed, and a P value of less than 0.05 was considered statistically significant.
Results. Mutant band in addition to wild bands were found in 6 (15.4%) of 39 pediatric AML patients, whereas 33 had only the NRAS wild-type allele (NRASwild). No apparent clinical or biologic characteristics were significantly different for the patients with NRAS gene mutations when compared with those without NRAS mutations except that there was lower peripheral and bone marrow blast (P=0.01, P=0.04) in patients with NRAS mutation when compared with those without mutation (Table 1).
Patients were assigned to the following cytogenetic groups: t(8;21) (15.4%; 6/39), inv(16) (15.4%6/39),del 5 (5.1%;2/39), del 7 (2.6%;1/39); and with CN-AML (53.8%; 21/39). In 7.7% (3/39) of the cases, conventional karyotyping was not available (Table 2). Patients with Inv(16) is presented with highest WBC count(median 41 x103/µL), and t(8;21)AML patients with lowest WBCs (median 18 x103/µL) compared with the other cytogenetic groups (Table 2). The median ages of children with Inv (16) AML (7 years), with t(8;21) (8.3 years), with del(7) (8.5), and with del 5(8.9 years) were younger compared with CN-AML (9.5 years).
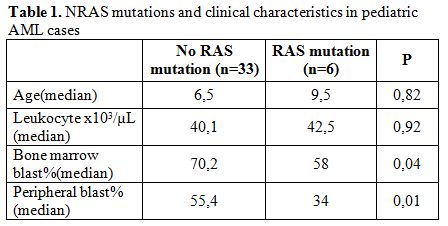
Table 1. NRAS mutations and clinical characteristics in pediatric AML cases.
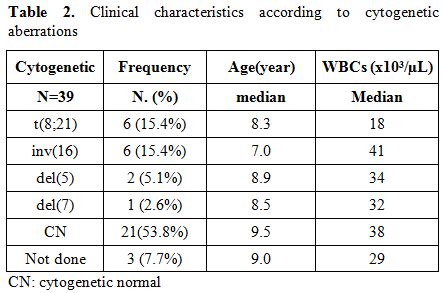
Table 2. Clinical characteristics according to cytogenetic aberrations.
In the FAB subtype M4e, NRAS mutations were represented more frequently (50%, 3 of 6) than they were in all other subtypes. In the M3, M5, M6 and M7 subtypes, no NRAS mutation was detected, making NRAS mutations highly underrepresented in these subtypes. In all other FAB subtypes, the distribution of NRAS mutations did not differ significantly from each other. A detailed distribution of NRAS mutations in the respective FAB subtypes is presented in Table 3.
Based on cytogenetic findings, the 39 patients were segregated into three groups: a good-risk group (n =12) was defined by karyotype, t(8;21) or inv(16); a poor-risk group (n =3) by del(5) or del(7) and standard-risk group (n =21) by normal karyotypes.
Based on the previous findings, we analyzed the influence of NRAS mutations on the prognosis of pediatric AML patients for whom clinical follow up data were available. Table 4 shows the clinical outcome in pediatric AML patients. In the total group, there was no difference with regard to CR rate (NRASmutant, 66.7%; NRASwild, 69.5%; P = 0.43). Relapse was significantly more frequent in the AML patients with NRAS gene mutations (NRASmuant, 33.4 %; NRASwild, 30.2 %; P= 0.26).
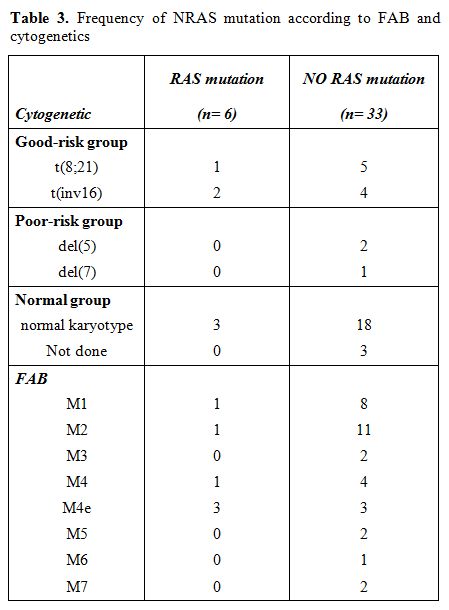
Table 3. Frequency of NRAS mutation according to FAB and cytogenetics.
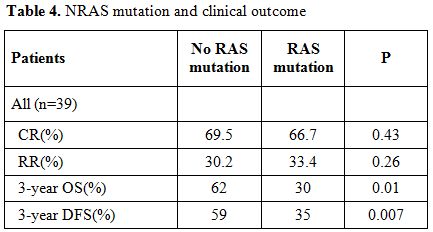
Table 4. NRAS mutation and clinical outcome.
At 3 years, the estimated OS was 30±15% in the presence of an NRAS mutation and 62±5% in its absence (P= 0.01). Adverse prognosis associated with N-ras mutations was observed in terms of 3 years DFS (NRASmutant 35% vs NRASwild 59%, P= 0.007), (Figure 2 and 3).
Univariate analysis showed that unfavorable prognostic factors for DFS were cytogenetic data (P = 0.005) and NRAS mutation (P = 0.002) (Table 5). Multivariate analysis showed NRAS was the strongest unfavorable factor (relative risk [RR], 3.6; P = 0.007), followed by cytogenetics (P = 0.02).
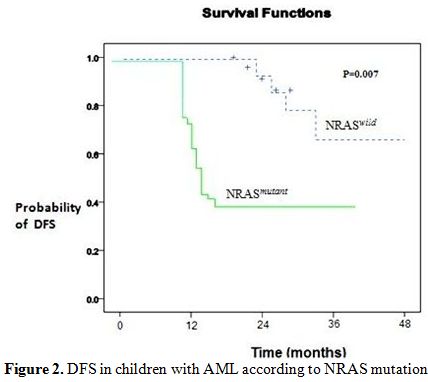
Figure 2. DFS in children with AML according to NRAS mutation.
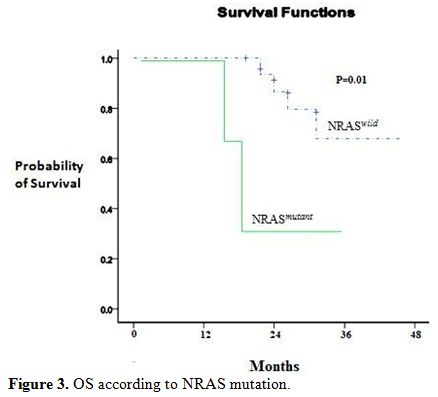
Figure 3. OS according to NRAS mutation.
Discussion
In recent years, a major focus of molecular cancer research has been the analysis of genes that may be causative in carcinogenesis (oncogenes). The clinical significance of RAS mutations has not been uniformly established. In the current study, we evaluated the clinical significance of NRAS mutations and investigated NRASmutant by genomic PCR method in 39 newly diagnosed pediatric AML cases.
Activated RAS mutations confer proliferative and survival signals. Mutations in the NRAS gene are frequent genetic aberrations in adult AML.[19] However, there have been only a few studies on childhood AML.[20] With different mutation-detection techniques used and heterogeneous patient populations studied, the reported incidence of NRASmutant in patients with childhood AML at presentation vary considerably; in our study 15.4% of pediatric AML patients (6/39) had NRASmutant, corresponding to the reported frequency by others.[15,21] Primary analyses revealed a statistically significant association between peripheral and bone marrow blast counts and NRAS mutation (P=0.01,P=0.04 respectively), however no significant differences had been found between the two groups with respect to age, gender, platelet count and WBCs count. These findings are in agreement with those reported in literatures.[10,13]
The highest frequency (33.3%) of NRAS mutations in our cohort (2/6) compared with the total cohort was detected in patients with inv(16). The high incidence of NRAS mutations in inv(16) in our study corresponded with most of the previously published studies reporting frequencies of 26% to 33% (22,23). 11q23/MLL aberrations are a frequent abnormality in pediatric AML.[24,25,26] The frequency of 11q23/MLL-rearranged AML may have been underestimated because of low number of cases in the included study and because in our study as well as in other studies performed in the past, the cryptic MLL rearrangements may be not detected by conventional karyotyping. It is conceivable that the biological differences may lead to different treatment strategies for these age categories in the future.[27] In our study, oldest children with AML were characterized by a high frequency of normal cytogenetic (53.8%) but the very young in the included study are characterized by higher frequency of inv (16).
The prognosis of AML depends on factors such as age, initial leukocyte count, FAB classification, karyotype, immune phenotype, and response to remission-induction therapy.[28,29] Our study showed that cytogenetic was unfavorable prognostic factor among AML patients by univariate analysis. This was in agreement with other study who found that cytogenetic data is thought to be the most important prognostic factor for AML.[30]
The prognostic significance of NRAS mutation in both adults and children remains disputed. Generally NRAS gene mutation is associated with tumor progression and was reported to be associated with poor prognosis in solid tumors and acute lymphoblastic leukemia (ALL).[31,32] Published reports addressing the clinical significance of NRAS mutations in patients with acute myeloid leukemia are inconclusive. Whereas some studies demonstrated a beneficial clinical effect of NRAS mutations,[33,34] others reached a different conclusion (e.g. lower CR).[35] Other studies also did not that show that patients with NRAS mutations had significantly good outcomes.[36,37]
In this study, the presence of NRAS gene mutation was related to similar complete remission (CR) rates following induction chemotherapy compared with non-mutated patients (66.7% vs. 69.5%, P=0.43). Those in CR had a similar relapse rate regardless of the presence of NRAS mutations (RR 33.4% vs. 30.2%, P=0.26). However, the presence of NRAS mutations was associated with poor three years OS and DFS compared with wild type cases (OS, P=0.01; DFS, P=0.007). This discrepancy between these studies findings and our study may be explained by differences in the intensity of the chemotherapy protocols employed to treat this group of patients and the small number of our cases.
Conclusions
In addition to the evidence that activation of the RAS-signaling cascade contributes to the molecular pathogenesis of myeloproliferative disorders (38), NRAS mutation has adverse prognostic impact but further pediatric studies will be necessary to extend our knowledge and more precisely define the prognostic significance of NRAS mutations. This study also demonstrates the need to screen for specific translocation partners to allow appropriate treatment stratification like WT1 and FLT3.
Acute myeloid leukemia (AML) is characterized by expansion of myeloid blasts with suppression of normal hematopoiesis. Cytogenetic and molecular studies have defined AML as a heterogeneous disease.[1] The presence of defined karyotypes is among the most important prognostic factors in acute myeloid leukemia (AML). However, even within defined cytogenetic groups stability of remission and long-term survival may vary significantly. Therefore, additional recurrent aberrations may have a prognostic impact.[2] It has been showed that Pediatric AML patients may harbor more than one mutation at diagnosis, some of which with a possible prognostic impact.[3-7] Mutations in the NRAS gene are one of these genetic aberrations that play a role in myeloid neoplasia.[8]
NRAS gene plays an important role in the regulatory processes that govern proliferation, differentiation and apoptosis; [9] abnormality in this gene has been implicated in the pathogenesis of AML. RAS oncogenes encode a family of membrane-associated proteins, which regulate signal transduction upon binding to a variety of membrane receptors.[10] There are three functional RAS genes (NRAS, KRAS and HRAS); in AML. K-RAS mutation occurs to a lower but still significant frequence in pediatric AML.[11] NRAS is the most prominent; reported in 11%-30% of patients.[12] All homologs were exclusively in codons [12,13], and 61 conferring constitutive activation of the RAS protein, which is subsequently held in the GTP bound status leading to an increased activity of the RAS pathway causing an increased proliferation and a decreased apoptosis rate.[13] RAS mutations were described in the various solid tumors as well as in hematologic malignancies. The prognostic impact of NRAS mutations is still under research and seems to vary from disease to disease,14 several studies indicated a poor prognostic impact for that mutation,[14,15] and however Lapillonne et al confirmed this finding in pediatric AML.[16] On the contrary, Neubauer et al found a favorable outcome for malignancies with NRAS mutations,12 and some studies failed to define any prognostic impact for NRAS mutations.[10,13,17]
We undertook this study to determine the frequency of NRAS mutation and its prognostic significance in a number of Egyptian pediatric patients with AML.
Patients and Methods
Newly diagnosed pediatric AML patients were included in this study; cases were recruited from pediatric hematology clinics of Tanta University Hospitals, Tanta, Egypt. Peripheral blood and BM samples were obtained from 39 de novo AML cases at initial diagnosis obtaining an informed consent from patients or their guardians; they were 21 boys and 18 girls. The median age was 7.4 years (range, 5.6-13 years). The median percentage of blasts in the fresh bone marrow samples was (65%). All included patients were receiving the same treatment protocol approved by the Oncology Team of Tanta University Hospital TUH; in brief they received 1-2 cycles of 14-21 days of intensively timed induction chemotherapy (doxorubicin, Ara-C, 6-thioguanine and methotrexate) depending upon BM aspiration done at the end of each induction course. Additional consolidation regimens included 1-2 cycles of 12-days (doxorubicin, Ara-C, VP-16 and methotrexate). Then patients who did not achieve remission received intermittent chemotherapy (Ara-C, 6-thioguanine and methotrexate) every 3 months for 6 cycles with the standard follow-up care and regular BM aspiration every 21 days to confirm remission; complete remission means normocellular bone marrow contain less than 5% blast cells and showing evidence of normal maturation of other bone marrow elements as evidenced by the repeated BM aspiration. The average duration of follow up was mean ± SD (32 ± 2.24 months).
Patients were classified according to the standard methods; morphological according FAB classification, cytochemical and immunological evaluation.[13] Informed consent was obtained from twenty subjects with matched age and sex who were selected as a control group. Samples from patients and control were analyzed for mutation in Exons 1, 2 of the NRAS gene using genomic PCR method.
Cytogenetic Analysis
Cytogenetic investigations were performed by karyotyping G-banding analysis in all patients.[18]
PCR of NRAS Gene
Genomic DNA was extracted from diagnostic bone marrow specimens of patients and control using the QIAamp DNA blood mini kit for DNA extraction provided by QIAGEN (Inc Chasworthy, CA). The concentration of extracted DNA was then measured by UV spectrophotometry at 260 & 280 nm and analyzed by electrophoresis on 2% agarose gel for detection of purity.
Separate assays were developed for mutation detection at (hot spots) in codons 12/13 (exon 1) and codon 61 (exon 2). Oligonucleotide primers amplifying short fragments {241 base pair (bp) for exon 1 and 201 bp for exon 2} were designed for PCR as follows:
N12/13 assay. forward, 5'-GACTGAGTACAAACTGGTGG-3'; and reverse, 5'-TGCATAACTGAATGTATACCC-3'.
N61 assay. forward, 5' CAAGTGGTTATAGATGGTGAAACC-3'; and reverse, 5'-AAGATCATCCTTTCAGAGAAAATAAT-3'.
PCR amplification was performed in 50 ul reaction, contained 50-100 ng of genomic DNA, 10 mM Tris HCl (pH 8.3), 50 mM KCl and 1.5 mM Mg Cl2, 200 uM of each deoxyribonucleotide triphosphate (dNTP), 2.5 units Taq polymerase, and 6% dimethylsulphoxide. PCR conditions were as follows: (1) Exon 1, HotStarTaq (Qiagen, Valencia, CA), and 0.625 U. Primers (12.5 pmol), N12/13 forward, N12/13 reverse. Denaturing, 95 C for 15 minutes and 94°C for 30 seconds; annealing, 55.5°C for 1minute; extension, 72°C for 1 minute for 35 cycles; final cycle at 72°C for 10 minutes. (2) Exon 2, HotStarTaq (Qiagen), 0.625 U. Primers (12.5 pmol), N61F forward, N61R reverse. Denaturing, 95°C for 15 minutes; 94°C for 30 seconds; annealing, 55.5°C for 1 minute; extension, 72°C for 1 minute for 35 cycles; final cycle at 72°C for 10 minutes.
Single-strand conformation polymorphism (SSCP) was performed to PCR product to detect mutations. Products were mixed with 10 volumes of loading buffer, quenched on ice immediately, and applied to 5% polyacrylamide gel electrophoresis at 50 V, overnight, stained by silver nitrates and wrapped in plastic foil. Normal gene exhibits a specific conformational pattern, while mutant gene displays pattern with different electrophoretic mobility (mobility shift) which was confirmed by repeated SSCP (Figure 1).

Figure 1. SSCP of PCR products from NRAS exon 1. C, normal marrow control, 1-6, samples from different pediatric patients. High molecular weight bands in lines 3 and 5 represent a normal NRAS band and a mutant band. Lines 1, 2, 4 and 6 represent wild type NRAS.
Statistical Methods
Data were processed and analyzed using SPSS for windows version 16.0 (SPSS, Inc, Chicago, IL, USA). Qualitative data were expressed as frequency and percentage and quantitative data were expressed as median. Chi-square test was used for comparative analysis. The prevalence of NRAS mutations in AML was too low to permit statistical analysis for correlation with survival. Kaplan-Meier analysis was used for survival of patients. The prognostic significance of the clinical variables was assessed using the Cox proportional hazards model. For all analyses, the P values were two-tailed, and a P value of less than 0.05 was considered statistically significant.
Results. Mutant band in addition to wild bands were found in 6 (15.4%) of 39 pediatric AML patients, whereas 33 had only the NRAS wild-type allele (NRASwild). No apparent clinical or biologic characteristics were significantly different for the patients with NRAS gene mutations when compared with those without NRAS mutations except that there was lower peripheral and bone marrow blast (P=0.01, P=0.04) in patients with NRAS mutation when compared with those without mutation (Table 1).
Patients were assigned to the following cytogenetic groups: t(8;21) (15.4%; 6/39), inv(16) (15.4%6/39),del 5 (5.1%;2/39), del 7 (2.6%;1/39); and with CN-AML (53.8%; 21/39). In 7.7% (3/39) of the cases, conventional karyotyping was not available (Table 2). Patients with Inv(16) is presented with highest WBC count(median 41 x103/µL), and t(8;21)AML patients with lowest WBCs (median 18 x103/µL) compared with the other cytogenetic groups (Table 2). The median ages of children with Inv (16) AML (7 years), with t(8;21) (8.3 years), with del(7) (8.5), and with del 5(8.9 years) were younger compared with CN-AML (9.5 years).

Table 1. NRAS mutations and clinical characteristics in pediatric AML cases.

Table 2. Clinical characteristics according to cytogenetic aberrations.
In the FAB subtype M4e, NRAS mutations were represented more frequently (50%, 3 of 6) than they were in all other subtypes. In the M3, M5, M6 and M7 subtypes, no NRAS mutation was detected, making NRAS mutations highly underrepresented in these subtypes. In all other FAB subtypes, the distribution of NRAS mutations did not differ significantly from each other. A detailed distribution of NRAS mutations in the respective FAB subtypes is presented in Table 3.
Based on cytogenetic findings, the 39 patients were segregated into three groups: a good-risk group (n =12) was defined by karyotype, t(8;21) or inv(16); a poor-risk group (n =3) by del(5) or del(7) and standard-risk group (n =21) by normal karyotypes.
Based on the previous findings, we analyzed the influence of NRAS mutations on the prognosis of pediatric AML patients for whom clinical follow up data were available. Table 4 shows the clinical outcome in pediatric AML patients. In the total group, there was no difference with regard to CR rate (NRASmutant, 66.7%; NRASwild, 69.5%; P = 0.43). Relapse was significantly more frequent in the AML patients with NRAS gene mutations (NRASmuant, 33.4 %; NRASwild, 30.2 %; P= 0.26).

Table 3. Frequency of NRAS mutation according to FAB and cytogenetics.

Table 4. NRAS mutation and clinical outcome.
At 3 years, the estimated OS was 30±15% in the presence of an NRAS mutation and 62±5% in its absence (P= 0.01). Adverse prognosis associated with N-ras mutations was observed in terms of 3 years DFS (NRASmutant 35% vs NRASwild 59%, P= 0.007), (Figure 2 and 3).
Univariate analysis showed that unfavorable prognostic factors for DFS were cytogenetic data (P = 0.005) and NRAS mutation (P = 0.002) (Table 5). Multivariate analysis showed NRAS was the strongest unfavorable factor (relative risk [RR], 3.6; P = 0.007), followed by cytogenetics (P = 0.02).

Figure 2. DFS in children with AML according to NRAS mutation.

Figure 3. OS according to NRAS mutation.
Discussion
In recent years, a major focus of molecular cancer research has been the analysis of genes that may be causative in carcinogenesis (oncogenes). The clinical significance of RAS mutations has not been uniformly established. In the current study, we evaluated the clinical significance of NRAS mutations and investigated NRASmutant by genomic PCR method in 39 newly diagnosed pediatric AML cases.
Activated RAS mutations confer proliferative and survival signals. Mutations in the NRAS gene are frequent genetic aberrations in adult AML.[19] However, there have been only a few studies on childhood AML.[20] With different mutation-detection techniques used and heterogeneous patient populations studied, the reported incidence of NRASmutant in patients with childhood AML at presentation vary considerably; in our study 15.4% of pediatric AML patients (6/39) had NRASmutant, corresponding to the reported frequency by others.[15,21] Primary analyses revealed a statistically significant association between peripheral and bone marrow blast counts and NRAS mutation (P=0.01,P=0.04 respectively), however no significant differences had been found between the two groups with respect to age, gender, platelet count and WBCs count. These findings are in agreement with those reported in literatures.[10,13]
The highest frequency (33.3%) of NRAS mutations in our cohort (2/6) compared with the total cohort was detected in patients with inv(16). The high incidence of NRAS mutations in inv(16) in our study corresponded with most of the previously published studies reporting frequencies of 26% to 33% (22,23). 11q23/MLL aberrations are a frequent abnormality in pediatric AML.[24,25,26] The frequency of 11q23/MLL-rearranged AML may have been underestimated because of low number of cases in the included study and because in our study as well as in other studies performed in the past, the cryptic MLL rearrangements may be not detected by conventional karyotyping. It is conceivable that the biological differences may lead to different treatment strategies for these age categories in the future.[27] In our study, oldest children with AML were characterized by a high frequency of normal cytogenetic (53.8%) but the very young in the included study are characterized by higher frequency of inv (16).
The prognosis of AML depends on factors such as age, initial leukocyte count, FAB classification, karyotype, immune phenotype, and response to remission-induction therapy.[28,29] Our study showed that cytogenetic was unfavorable prognostic factor among AML patients by univariate analysis. This was in agreement with other study who found that cytogenetic data is thought to be the most important prognostic factor for AML.[30]
The prognostic significance of NRAS mutation in both adults and children remains disputed. Generally NRAS gene mutation is associated with tumor progression and was reported to be associated with poor prognosis in solid tumors and acute lymphoblastic leukemia (ALL).[31,32] Published reports addressing the clinical significance of NRAS mutations in patients with acute myeloid leukemia are inconclusive. Whereas some studies demonstrated a beneficial clinical effect of NRAS mutations,[33,34] others reached a different conclusion (e.g. lower CR).[35] Other studies also did not that show that patients with NRAS mutations had significantly good outcomes.[36,37]
In this study, the presence of NRAS gene mutation was related to similar complete remission (CR) rates following induction chemotherapy compared with non-mutated patients (66.7% vs. 69.5%, P=0.43). Those in CR had a similar relapse rate regardless of the presence of NRAS mutations (RR 33.4% vs. 30.2%, P=0.26). However, the presence of NRAS mutations was associated with poor three years OS and DFS compared with wild type cases (OS, P=0.01; DFS, P=0.007). This discrepancy between these studies findings and our study may be explained by differences in the intensity of the chemotherapy protocols employed to treat this group of patients and the small number of our cases.
Conclusions
In addition to the evidence that activation of the RAS-signaling cascade contributes to the molecular pathogenesis of myeloproliferative disorders (38), NRAS mutation has adverse prognostic impact but further pediatric studies will be necessary to extend our knowledge and more precisely define the prognostic significance of NRAS mutations. This study also demonstrates the need to screen for specific translocation partners to allow appropriate treatment stratification like WT1 and FLT3.
References
- DG, Griffin JD. The roles of FLT3 in
hematopoiesis and leukemia. Blood 2002; 100: 1532–1542. http://dx.doi.org/10.1182/blood-2002-02-0492
PMid:12176867

- Ritter M, Kim TD, Lisske P, Thiede C,
Schaich M, Neubauer A. Prognostic significance of N-RAS and K-RAS
mutations in 232 patients with acute myeloid leukemia. Haematologica
2004 89: 1397-1399. PMid:15531466

- Bachas C, Schuurhuis GJ, Hollink IH,
Kwidama ZJ, Goemans BF, Zwaan CM, van den Heuvel-Eibrink MM, de Bont
ES, Reinhardt D, Creutzig U, de Haas V, Assaraf YG, Kaspers GJ, Cloos
J. High-frequency type I/II mutational shifts between diagnosis and
relapse are associated with outcome in pediatric AML: implications for
personalized medicine. Blood. 2010 Oct 14;116(15):2752-8. Epub 2010 http://dx.doi.org/10.1182/blood-2010-03-276519
PMid:20592250

- Meshinchi S, Woods WG, Stirewalt DL,
Sweetser DA, Buckley JD, Tjoa TK, Bernstein ID, Radich JP.Prevalence
and prognostic significance of Flt3 internal tandem duplication in
pediatric acute myeloid leukemia. Blood. 2001 Jan 1;97(1):89-94 http://dx.doi.org/10.1182/blood.V97.1.89
PMid:11133746

- Meshinchi S, Stirewalt DL, Alonzo TA, Zhang
Q, Sweetser DA, Woods WG, Bernstein ID, Arceci RJ, Radich JP.
Activating mutations of RTK/ras signal transduction pathway in
pediatric acute myeloid leukemia. Blood. 2003 Aug 15;102(4):1474-9. http://dx.doi.org/10.1182/blood-2003-01-0137
PMid:12702504

- Hollink IH, van den Heuvel-Eibrink MM,
Zimmermann M, Balgobind BV, Arentsen-Peters ST, Alders M, Willasch A,
Kaspers GJ, Trka J, Baruchel A, de Graaf SS, Creutzig U, Pieters R,
Reinhardt D, Zwaan CM.Clinical relevance of Wilms tumor 1 gene
mutations in childhood acute myeloid leukemia. Blood. 2009 Jun 4;113
(23):5951-60. http://dx.doi.org/10.1182/blood-2008-09-177949
PMid:19171881

- Ho PA, Zeng R, Alonzo TA, Gerbing RB,
Miller KL, Pollard JA, Stirewalt DL, Heerema NA, Raimondi SC, Hirsch B,
Franklin JL, Lange B, Meshinchi S.Prevalence and prognostic
implications of WT1 mutations in pediatric acute myeloid leukemia
(AML): a report from the Children's Oncology Group. Blood. 2010 Aug
5;116 (5):702-10, http://dx.doi.org/10.1182/blood-2010-02-268953
PMid:20413658 PMCid:2918327

- Lubbert M, Mirro J Jr., Kitchingman G, et
al. Prevalence of N-ras mutations in children with myelodysplastic
syndromes and acute myeloid leukemia. Oncogene. 1992;7:263–268.
PMid:1549347

- Hirai H. Molecular Mechanisms of
Myelodysplastic Syndrome. Jpn. J. Clin. Oncol 2003;33 (4): 153-160. http://dx.doi.org/10.1093/jjco/hyg037
PMid:12810828

- Bowen D, Frew M, Hills R et al. RAS
mutation in acute myeloid leukemia is associated with distinct
cytogenetic subgroups but does not influence outcome in patients
younger than 60 years. Blood 2005; 106: 2113-2119. http://dx.doi.org/10.1182/blood-2005-03-0867
PMid:15951308

- Goemans BF, Zwaan CM, Miller M, Zimmermann
M, Harlow A, Meshinchi S, Loonen AH, Hählen K, Reinhardt D, Creutzig U,
Kaspers GJ, Heinrich MC. Mutations in KIT and RAS are frequent events
in pediatric core-binding factor acute myeloid leukemia. Leukemia. 2005
Sep;19(9):1536-42. http://dx.doi.org/10.1038/sj.leu.2403870
PMid:16015387

- Beaupre DM, Kurzrock R. RAS and leukemia:
from basic mechanisms to gene-directed therapy. J Clin Oncol.
1999;17:1071-1079. PMid:10071302

- Illmer T, Thiede C, Fredersdorf A et al.
Activation of the RAS pathway is predictive for a chemosensitive
phenotype of acute myelogenous leukemia blasts. Clin Cancer Res 2005;
11: 3217-3224. http://dx.doi.org/10.1158/1078-0432.CCR-04-2232
PMid:15867216

- Padua RA, West RR. Oncogene mutation and
prognosis in the myelodysplastic syndromes. Br J Haematol 2000;
111:873-874. http://dx.doi.org/10.1046/j.1365-2141.2000.02472.x
PMid:11122149

- Paquette RL, Landaw EM, Pierre RV et al.
N-Ras mutations are associated with poor prognosis and increased risk
of leukemia in myelodysplastic syndrome. Blood 1993;82: 590-599
PMid:8329714

- Lapillonne H, Liopis L, Auvrignon A,
Renneville A, Labopin M, Mazingue F, Perot C, et al. Extensive
mutational status of genes and clinical outcome in pediatric acute
myeloid leukemia. Leukemia 2010; 24, 205–209. http://dx.doi.org/10.1038/leu.2009.172
PMid:19759559

- Radich JP, Kopecky KJ, Willman CL et al.
N-Ras mutations in adult de novo acute myelogenous leukemia: prevalence
and clinical significance. Blood 1990; 76:801-807. PMid:2200539

- Mitelman F (ed). (1991). An international system for human cytogenetic nomenclature. Karger: Basel,1991.
- Schaich M, Ritter M, Illmer T, et al.
Mutations in ras proto-oncogenes are associated with lower mdr1 gene
expression in adult acute myeloid leukaemia. Br J Haematol 2001; 112:
300-307. http://dx.doi.org/10.1046/j.1365-2141.2001.02562.x
PMid:11167822

- Vogelstein B, Civin CI, Preisinger AC, et
al. RAS gene mutations in childhood acute myeloid leukemia: a Pediatric
Oncology Group study. Genes Chromosomes Cancer 1990; 2: 159-162. http://dx.doi.org/10.1002/gcc.2870020212

- Liang DC, Shih LY, Fu JF, et al. K-ras
mutations and N-ras mutations in childhood acute leukemias with or
without mixed-lineage leukemia gene rearrangements. Cancer 2006; 106:
950-956. http://dx.doi.org/10.1002/cncr.21687
PMid:16404744

- Valk PJM, Bowen DT, Frew ME, et al. Second
hit mutations in the RTK/RAS signaling pathway in acute myeloid
leukemia with inv(16) [abstract]. Haematologica 2004; 89:106.
PMid:14754614

- Beghini A, Peterlongo P, Ripamonti CB, et
al. C-kit mutation in core binding factor leukemias. Blood 2000;
95:726-727. PMid:10660321

- Harrison CJ, Hills RK, Moorman AV,
Grimwade DJ, Hann I, Webb DK, Wheatley K, de Graaf SS, van den Berg E,
Burnett AK, Gibson BE. Cytogenetics of childhood acute myeloid
leukemia: United Kingdom Medical Research Council Treatment trials AML
10 and 12. J Clin Oncol. 2010 Jun 1;28(16):2674-81. Epub 2010http://dx.doi.org/10.1200/JCO.2009.24.8997
PMid:20439644

- von Neuhoff C, Reinhardt D, Sander A,
Zimmermann M, Bradtke J, Betts DR, Zemanova Z, Stary J, Bourquin JP,
Haas OA, Dworzak MN, Creutzig U. Prognostic impact of specific
chromosomal aberrations in a large group of pediatric patients with
acute myeloid leukemia treated uniformly according to trial AML-BFM 98.
J Clin Oncol. 2010 Jun 1;28(16):2682-9. http://dx.doi.org/10.1200/JCO.2009.25.6321
PMid:20439630

- Bachas C, Schuurhuis GJ, Hollink IH,
Kwidama ZJ, Goemans BF, Zwaan CM, van den Heuvel-Eibrink MM, de Bont
ES, Reinhardt D, Creutzig U, de Haas V, Assaraf YG, Kaspers GJ, Cloos
J. High-frequency type I/II mutational shifts between diagnosis and
relapse are associated with outcome in pediatric AML: implications for
personalized medicine. Blood. 2010 Oct 14;116(15):2752-8. http://dx.doi.org/10.1182/blood-2010-03-276519
PMid:20592250

- Balgobind BV, Hollink IH, Arentsen-Peters
ST, Zimmermann M, Harbott J, Beverloo B, von Bergh AR, Cloos J, Kaspers
GJ, de Haas V, Zemanova Z, Stary J, Cayuela JM, Baruchel A, Creutzig U,
Reinhardt D, Pieters R, Zwaan CM, van den Heuvel-Eibrink MM.Integrative
analysis of type-I and type-II aberrations underscores the genetic
heterogeneity of pediatric acute myeloid leukemia. Haematologica. 2011,
96(10):1478-1487 http://dx.doi.org/10.3324/haematol.2010.038976
PMid:21791472 PMCid:3186309

- Kobayashi T, Miyawaki S, Tanimoto M,
Kuriyama K, et al. Randomized trials between behenoyl cytarabine and
cytarabine in combination induction and consolidation therapy, and with
or without ubenimex after maintenance/ intensification therapy in adult
acute myeloid leukemia. The Japan Leukemia Study Group. J Clin Oncol
1996; 14:204. PMid:8558199

- Miyawaki S, Tanimoto M, Kobayashi T, Minami S, et al. Randomized study of individualized induction therapy with or without etoposide in adult acute myeloid leukemia (except for M3) (JALSG-AML92 study) and analysis of prognostic factors. Blood 1997; 90:503a.
- Mro’zek K, Heinonen K, de la Chapelle A,
Bloomfield CD: Clinical significance of cytogenetics in acute myeloid
leukemia. Semin Oncol 1997; 24:17. PMid:9045301

- Slebos RJ, Kibbelaar RE, Dalesio O,
Kooistra A, Stam J, Meijer CJ, Wagenaar SS, Vanderschueren RG, van
Zandwijk N, Mooi WJ, Bos JL, Rodenhuis S: K-RAS oncogene activation as
a prognostic marker in adenocarcinoma of the lung. N Engl J Med 1990;
323;9: 561-565. http://dx.doi.org/10.1056/NEJM199008303230902
PMid:2199829

- Lübbert M, Mirro J Jr, Miller CW, Kahan J,
Issac G, Kitchingman G, Mertelsmann R, Herrmann F, McCormick F,
Koeffler HP: N-Ras gene point mutations in childhood acute lymphocytic
leukemia correlate with a poor prognosis. Blood 1990; 75:1163-1169.
PMid:2407301

- Neubauer A, Dodge RK, George SL, Davey FR,
Silver RT, Schiffer CA, et al. Prognostic importance of mutations in
the ras proto-oncogenes in de novo acute myeloid leukemia. Blood
1994;83:1603-11. PMid:8123851

- Coghlan DW, Morley AA, Matthews JP, Bishop
JF. The incidence and prognostic significance of mutations in codon 13
of the Nras gene in acute myeloid leukemia. Leukemia 1994; :1682-7.
PMid:7934163

- Kiyoi H, Naoe T, Nakano Y, Yokota S,
Minami S, Miyawaki S, et al. Prognostic implication of FLT3 and N-RAS
gene mutations in acute myeloid leukemia. Blood 1999;93:3074-80.
PMid:10216104

- De Melo MB, Lorand-Metze I, Lima CS, Saad
ST, Costa FF N-ras gene point mutations in Brazilian acute myelogenous
leukemia patients correlate with a poor prognosis. Leuk Lymphoma.
1997;24 :309-17. PMid:9156660

- Stirewalt DL, Kopecky KJ, Meshinchi S,
Appelbaum FR, Slovak ML, Willman CL, et al. FLT3, RAS, and TP53
mutations in elderly patients with acute myeloid leukemia. Blood
2001;97:3589- 95. http://dx.doi.org/10.1182/blood.V97.11.3589
PMid:11369655

- Chan IT, Kutok JL, Williams IR, Cohen S,
Kelly L, Shigematsu H, et al. Conditional expression of oncogenic K-ras
from its endogenous promoter induces a myeloproliferative disease. J
Clin Invest 2004;113:528-38. PMid:14966562 PMCid:33826
