Received: November 4, 2013
Accepted: January 18, 2014
Meditter J Hematol Infect Dis 2014, 6(1): e2014015, DOI 10.4084/MJHID.2014.015
This article is available on PDF format at:

Mohammad Varahram, Parissa Farnia, Mohammad Javad Nasiri, Mona Afraei Karahrudi, Mehdi Kazempour Dizagie and Ali Akbar Velayati
Mycobacteriology
Research Centre, National Research Institute of Tuberculosis and Lung
Disease [NRITLD], Masih Daneshvari Hospital, Shahid Beheshti University
of Medical Sciences.

|
This
is an Open Access article distributed
under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0),
which permits unrestricted use, distribution, and reproduction in any
medium, provided the original work is properly cited.
|
|
Abstract The
six major lineages of Mycobacterium tuberculosis [MTB] are found to be
strongly associated with specific geographical outbreaks. But whether
these bacterial lineages influence the host genetic polymorphism is
uncertain. The present study was designed to evaluate the relevance of
strain diversity and host genetic polymorphisms in susceptibility to
pulmonary tuberculosis [PTB]. For this reason, single –nucleotide
polymorphisms [SNPs] in interferon- γ [IFN-γ] receptor-1[G-611A], IFNG
[G+ 2109A] and tumor necrosis factors [TNF-α] genes [at -238,
308,-857position] in patients [n=151] were analyzed and compared with
controls [n=83]. The genetic diversity of M. tuberculosis isolates was
performed using spacer oligonucleotide typing. Thereafter, the profile
of IFN-γ and TNF-α allele frequency were investigated in each subtype
of M. tuberculosis. The results showed C allele of TNF 857 and A allele
of TNF 238 were more frequent in PTB cases [[TNF 857 C allele OR
[CI95%] 0.6[0.4-0.9], p= 0.02] for TNF 238 A allele OR [CI95%]
5.5[3.4-9.0], p= 0.00]]. Similarly, G allele in IFNG+ 2109 A/G
polymorphism were significantly more in patients than control subject
[OR[CI95%] 0.3; p< 0.05]. The major identified clinical isolates
of
M. tuberculosis were EAI [42; 27.8%], Haarlem [31; 20.5%], CAS [23;
15.2%], Beijing [14; 9.2%], and T [11; 7.2%] lineages. No correction
was observed between strains diversity and frequency of SNPs in studied
PTB cases. In conclusions, we exclude the possibility of genetic
mutation in IFN-γ and TNF-α gene by different subtypes of M.
tuberculosis. Although, our results supports a positive correlation
between host SNPs and susceptibility to PTB. |
Introduction
Tuberculosis [TB], caused by Mycobacterium tuberculosis,
is a major cause of morbidity and mortality throughout the world. It is
estimated that one third of the world’s population is infected with M. tuberculosis,
and approximately 1 billion people will be added to this number till
2020.[1] Among those who are
infected, only 5-10% will develop the
active form of the disease with clinical symptoms. Other infected
individuals may remain noninfectious and symptoms free for many
years.[2] Basically, the course of
infection depends on a complex
interaction of host, bacteria and environmental factors.[3]
The genetic
contribution of the host in the individual susceptibility and
development of disease is well studied during recent years.[3] In this
regards, both genes for interferon-gamma [IFN-γ] and tumor necrosis
factor-alpha [TNF-α] have been identified as a essential components of
the host immune response.[4,5]
IFN-γ is the key cytokine involved in
the protective response against M.
tuberculosis infection
and is required for control of this pathogen. TNF-α in synergy with
IFN-γ induce antimycobacterial activity of macrophages and increases
its bactericidal activity.[6] Till
date, several polymorphisms within
the promoter region of TNF-α and IFN-γ gene have been shown to be
associated with susceptibility or resistance to TB in different ethnic
groups.[7-10] In contrast, the role
of genetic variability of M.
tuberculosis
in the outcome of the infection remains to be uncertain.[11] Most of
immunological research on tuberculosis has been performed with
laboratory strains i.e., H37RV and Erdman. But, with advances in
molecular biology, it became apparent that M. tuberculosis
is not a genetically conserved bacterium with limited phenotypic
differences.[11,12] Additionally,
studies on molecular epidemiology
showed differences in transmissibility and virulence of various
subtypes of M.
tuberculosis.
Lopez and his co-workers were among the first investigators who could
represent different immunopathological events using different M. tuberculosis
strains.[11] Later on, Tanveer et
al. studied the cytokine secretion in
patients that were infected by CAS1 and Beijing subtypes of M. tuberculosis.[12] In other studies, the influences of
M. tuberculosis
lineages to innate immune responses were characterized.[13,14]
However, association between host genetic polymorphisms and
susceptibility to different lineages of M. tuberculosis
strains was not reported. Recently, we showed the high prevalence of
Beijing and Haarlem lineages among Iranian drug resistance TB
patients.[7,15]
Initially, Beijing was described in 1995 as a closely
related group of tubercle bacilli from the People's Republic of China,
and Haarlem were mainly found in Central America and Caribbean.[16,17]
To date, the prevalence of Beijing and Haarlem have reported in several
countries.[16,18]
In the present study, the association of IFN-γ and
TNF-α polymorphisms with susceptibility to TB in genetically diverse
subtypes of M.
tuberculosis
are investigated. To our knowledge this is the first report that
investigates association of host genetic polymorphism with genotyping
of M. tuberculosis.
Material and Methods
Setting
and study population.
The study was conducted from January 2010 to December 2012, in the
Mycobacteriology Research Center [MRC]. MRC is the only WHO-approved
center for the detection and diagnosis of TB patients in Iran. A total
of 151 patients with culture-positive TB and 83 healthy volunteers
[referred to as normal controls] were included in the study. Patients
and control subjects were matched for age, sex and nationality [The
Institutional Review Board at the NRITLD approved the study and all the
patients have signed informed consent].
Mycobacterial
isolates.
Collected sputum samples from each patient were digested and
decontaminated by Petroff’s method.[19]
Lowenstein-Jensen media were
used for bacterial growth. The extracted DNA from culture positive
samples was used for identification and spoligotyping.[15,20] Drug
susceptibility testing was performed against first–line anti TB drugs
using proportional method.[21]
Spoligotyping
of MTB isolates.
Spoligotyping was performed for all 151 clinical isolates according to
the standard method.[20] Briefly,
DR region of mycobacterial genome was
amplified by PCR using following primers: DRa 5' - GGT TTT GGG TCT GAC
GAC -3' [biotinylated at 5'end] and DRb 5'-CCG AGA GGG GAC GGA AAC-3'
[Metabion, Martinsried, Germany]. The PCR amplicons were subsequently
hybridized to a set of 43 different immobilized DR spacers covalently
bound to the membrane. The hybridization signals were detected by
chemiluminescence system [Amersham ECL detection kit, GE Healthcare
Limited, UK] after incubation with a streptavidin-peroxidase conjugate
[Roche, Germany]. DNA extracts of MTB H37Rv and M. bovis BCG were
used as positive controls.
Genetic
evaluation.
Genomic DNA was extracted using the standard protocol with slight
modifications.[23,24] Briefly,
Peripheral Blood Leukocytes [PBLs] were
separated from two milliliters of the whole blood using RBC lysis
buffer [0.155 M NH4Cl,
0.01 M NaHCO3].
Thereafter, PBLs re-suspended in
500µl of SE buffer [NaCl 3M, EDTA 0.5M, PH=8], containing 40 µl of 10%
SDS and 3µl of 20 mg/ml of proteinase K. The suspension was incubated
at 60°C for 30 minutes. After incubation, 200µl of equilibrated phenol
[PH=8] was added to the mixture and centrifuged for 10 min at 12000g.
The aqueous phase transferred to a new tube and the DNA was
precipitated using cold propanol.
TNF-α
genotyping.
Polymorphisms in the TNF promoter region, namely TNF single nucleotide
polymorphisms [SNP] 238, 308 and 857 were studied using PCR- RFLP. For
TNF –308 polymorphisms, the following primers were used to amplify a
107bp product:5' AGC AAT AGG TGG TTT TGA CTC GGGC CCAT-3';5'TCC TCC CTG
CTC CGA TTC CG-3'. For -238 polymorphisms, the following primers were
used to amplify a 230 bp product : 5'CCT CAA GGA CTC CAA AGC TTT CTG
-3'; 5'ACA CTC CCC ATC CTC CCA GATC -3'. For -857 polymorphisms, the
following primers were used to amplify a 127 bp product: 5' GGC TCT GAG
GAA TGG GTT AC-3' ;5'CCT CTA CAT GGC CCT GTC TAC-3'. The amplification
was accomplished by an initial denaturation at 94oC for 5 min, and 30
cycles at 94oC
for 40s, at 56oC
for 40s, at 72oC
for 1 min, followed by
an extension at 72oC for 6 min.[22,24] PCR products of, TNF -238, TNF
-308 and TNF -857 digested with 2 U enzymes of BgI II, Bsaj I, NcoI,
TaiI and TaiI,respectively.[24]
Digested products were run on 8%
polyacrylamide gel and were stained with Silver –Nitrate.
IFN-γ
genotyping.
Single–nucleotide polymorphisms [SNPs] in interferon-γ [IFN-γ]
receptor-1[G-611A], IFNG [G+ 2109A] were performed using PCR-RFLP. The
target DNA was amplified in a PCR reaction mixture containing 1×
reaction buffer [50 mM KCl, 67 mM Tris-HCl [pH 9.0], 2 mM MgCl2], 2 mM
of dNTPs, 0.2U of Taq DNA-polymerase [Roch, Germany], and 20 pmol of
each primers. For IFN-γ receptor-1[G-611A s, the following primers were
used to amplify a 85bp product: 5' AGACAAACCCAGAGAGGTAAGAGA3';
5'ACCTTCTCAGCAATTCAGTGTCAAA3'. For IFNG [G+ 2109A] polymorphisms, the
following primers were used to amplify a 366 bp Product;
5'AATCGCTGAAGTATGTAAT3'; 5'GCATTGTAGAGTTTTG3'. The PCR products of
IFN-γ receptor-1[G-611A] and IFNG [G+ 2109A] digested with 2 U enzymes
of NIaIII and FauI, respectively.[24]
Digested products were run on 8%
polyacrylamide gel and were stained with Silver –Nitrate.
Statistical
analysis.
Statistical analysis was performed using chi-square test to determine
statistical associations between cases and control. P-value less than
0.05 were considered statistically significant. Data were analyzed
using SPSS version 18 Software.
Results
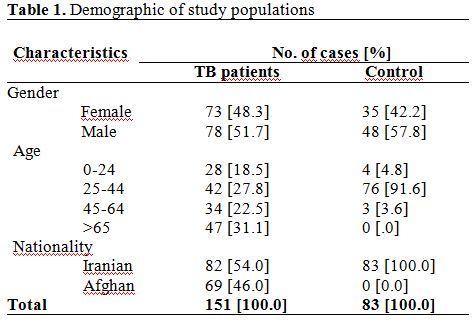
According to the demographic characteristics of studied populations,
the mean age of patient and control groups was 48.7±22.1 and 32.7±6.4
years, respectively (Table
1).
Majority of studied patient [47; 31.2%] had more than 65 years of old,
followed by second group [42; 27.8 %] which had 25-44 years of ages.
Seventy three [48.2%] TB cases were female and 78 [51.7%] were males;
while in the control group 34 [42.2%] were females and 48 [57.8%] were
males. Most of the patients [54.0%] were Iranian, and remains were
immigrants.
 | Table 1. Demographic of study populations |
Spoligotyping. Of 151 MTB isolates for which spoligotyping was performed, 140 [92.6%] isolates were grouped into 13 different “shared type” that had been described in the SITVIT2 database and the remaining 11 [7.4%] isolates generated unknown spoligopatterns. The most frequent spoligotype in our populations belong to EAI [EAI1 and EAI3, n=42, 27.8%], Haarlem [H3 and H4, n=31, 20.5%] followed by CAS [CAS1 and CAS2, n=23, 15.2%], Beijing [n=14, 9.2%], and T [T, T3 and T4, n=11; 7.2%] lineages (Table 2).
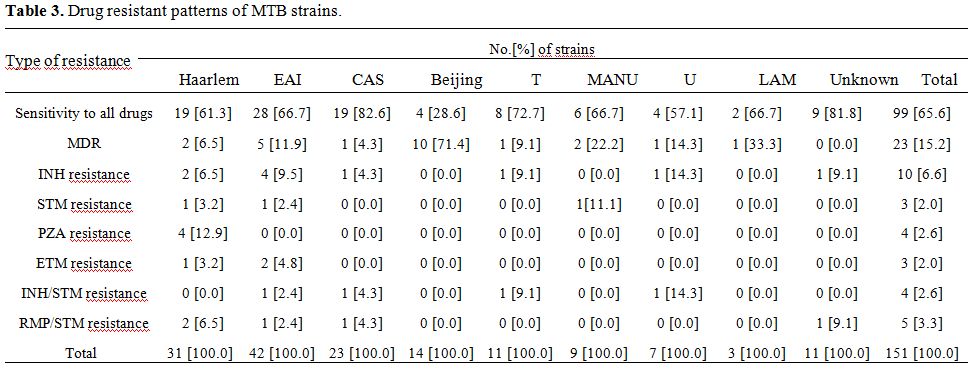
Drug susceptibility patterns. As shown in table 3, drug susceptibility testing of 151 strains indicated that 99 [65.6%] were sensitive to all tested agents and 52 [34.4%] were resistant to at least one drug. The majority of drug resistant isolates were resistance to INH [6.6%] followed by PZA [2.6%] and ETM [2.0%] and STM [2.0%]. None of the investigated isolated were RMP monoresistant. Twenty three isolates were MDR-TB [15.2%]. In an investigation between MTB strains and drug resistance we found that Beijing genotype was highly associated with MDR [p<0.05].
Association of IFN-γ and TNF-α gene polymorphisms with TB. Allele and the genotype frequencies of investigated IFN-γ and TNF-α polymorphisms are enlisted in Table 4. In overall, three types of polymorphisms were observed in TNF-α gene: an A to G substitution at position -238, a G to A substitution at position -308, and a C to T substitution at position -857. Among these polymorphisms, C allele of TNF 857 and A allele of TNF 238 were more frequent in TB cases as compared to control group [TNF 857 C allele OR[CI95%] 0.6[0.4-0.9], p= 0.02] for TNF 238 A allele [OR[CI95%] 5.5[3.4-9.0], p= 0.00]. Additionally, TNF 857 C/C[ 85;56.2%] and TNF 238 A/A 127[84.1%] genotypes were associated with increased risk of acquiring TB. Two types of polymorphisms, in an A to G substitution at position + 2109 and -611, were observed for IFN-γ. The result showed in - 2109 A/G polymorphism, G allele were significantly more common in TB group [OR[CI95%] 0.3; p< 0.05]. The investigation of the allele and genotype frequencies for TNF- 308 and IFNR1 -611 polymorphisms revealed no significant association with resistance or susceptibility to TB [p > 0.05; Table 4].
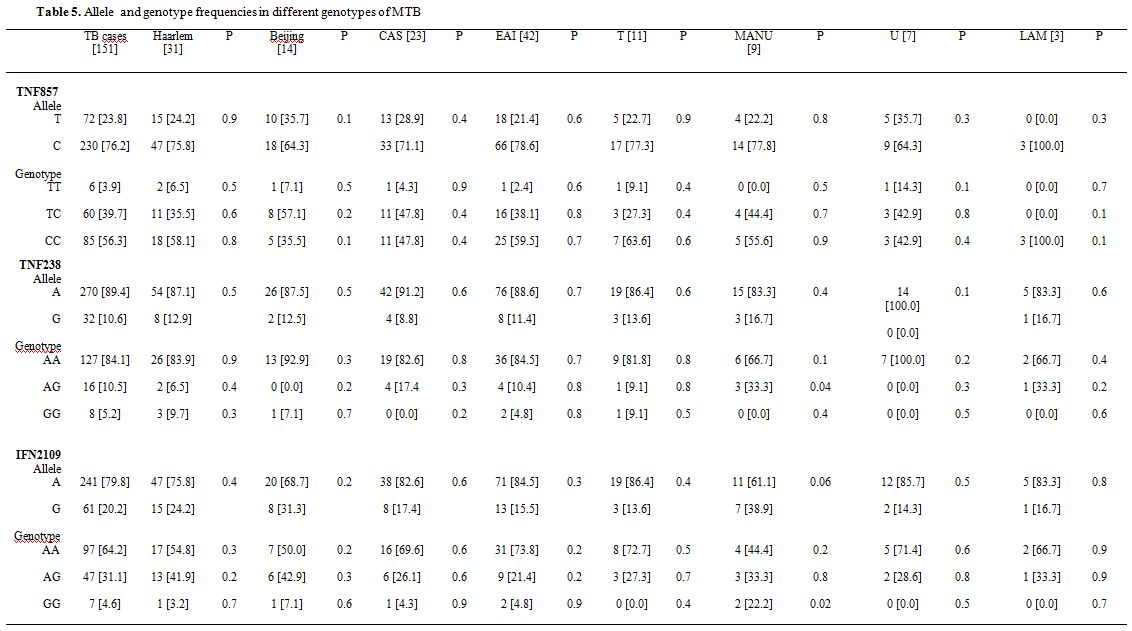
Association of IFN-γ and TNF-α polymorphisms with major lineages of M. tuberculosis. As shown in table 5, three polymorphic variants [two in TNF-α gene, and one in IFN-γ] are associated with susceptibility to TB. Distributions of these alleles are similar in patients that were infected with different subtypes of M. tuberculosis. For example, out of 42, 31, 23, 19, 14 TB patients infected with EAI, Haarlem, CAS, T, Beijing lineages, 88.6%, 87.1%, 91.2%, 86.4% and 87.5%, had TNF 238A allele, respectively. Similarly, the frequency of IFN-γ A allele was high in all TB patients [ranging from 61.1 to 86.4%]. Thereby, we found no correlation between host genetic polymorphisms and mycobacterial diversity.
 | Table 2. The spoligotyping pattern of MTB strains |
 | Table 3. Drug resistant patterns of MTB strains |
 | Table 4. Allele and genotype frequencies in TB cases |
 | Table 5. Allele and genotype frequencies in different genotypes of MTB |
Discussion
Pathogenesis in tuberculosis is dependent on many components of the host, pathogen and environment.[27] The present study was aimed to evaluate the possible correlation of host genetic polymorphisms with different genotypes of M. tuberculosis. Based on SIT from SITVIT2, the major identified clinical isolates of M. tuberculosis were EAI [42; 27.8%], Haarlem [31; 20.5%], CAS [23;15.2% ], Beijing [14; 9.2%], and T [11; 7.2%] lineages. Recently, it was shown that particular genotypes of M. tuberculosis could elicit different immune responses with high mortality rates in the course of experimental infection.[11-13] For example, in mice model Beijing subtypes, induced early and massive pneumonia with death. Whereas, Canetti strains induced limited pneumonia with sustained expression of TNF-α.[27,29] Likewise, other investigators showed a different level of cytokines production [TNF-α,IFN-γ] in patients infected with genetically distinct M. tuberculosis subtypes.[12] Our results showed no association between the frequencies of SNPs in host and various lineages of M. tuberculosis. As shown in table 5, the TNF-α 238A and 857C alleles were associated with susceptibility to TB infection, but their distribution was almost equal among patient infected with different subtypes of M. tuberculosis. Likewise, the frequency of IFN2109A allele was high in TB patients than control subject, but no statistical differences were observed among the allele distribution in different M. tuberculosis lineages. Therefore, our results demonstrate no correlation between genetic diversity of M. tuberculosis and host susceptibility to TB. On the contrary to our results, Tanveer et al. showed a correlation between cytokine induction i.e., TNF-α, IFN-γ and growth index of CAS1 and Beijing isolates in comparison to H37RV strain.[12] Furthermore, they suggested that the phenotypic and genotypic polymorphisms in clinical M. tuberculosis strains may in turn influence the persistence and dissemination of differing genotypes.
At present, we have no explanation for such discrepancy, but we need more detailed studies in order to outline the importance of M. tuberculosis genotypes with host genetic polymorphism. Basically, genetic contribution of the host is an important factor in determining susceptibility to TB. Today, several cytokine gene polymorphisms have been described in association with susceptibility or resistance to TB. In present investigations, we found two polymorphisms of TNF gene promoter [-857 and -238] that were significantly associated with TB patients (Table 4). For TNF- 238 A/G SNP, A allele was more frequent in TB cases as compared to control. Previously also, positive association of TNF- 238 A/G was reported among Iranian pulmonary tuberculosis cases.[22,30] TNF 238 A/G polymorphism has been extensively studied in TB cases of various ethnic groups.[8-10] However, studies that were conducted in Turkey, India and Columbia demonstrated no association of specific allele of TNF-α gene with susceptibility to TB.[8,9,31] There are also considerable variations in genotype frequencies of TNF 857 polymorphisms in different populations. TNF 857 T/C polymorphism in our study was significantly more frequent in TB cases as compared to control. However, conflicting reports are available about insignificant association of 857T/C genotype in Asian TB patients i.e. Indians.[4] In fact, the contradictory data could be discussed in different ways; First of all, multiple polymorphisms within the TNF gene may have emerged during evolution in various ethnic groups to affect TB susceptibility or resistance. Second, the number of studied cases has a great impact on the outcome of the results. Generally, the large confidence intervals in some studies could be the result of the small sample size. For TNF 308 G/A, several studies on TB patients have produced approximately similar results. In recent surveys, no significant association in TNF 308 and TB were reported from Korea, Brazil and China, which is similar to our study.[9,32,33]
Another candidate gene for determination the susceptibility to TB is polymorphism in the IFN-γ gene.[3,26] In different experimental set up, tuberculosis patients had deficient IFN-γ production in their peripheral blood mononuclear cells. Also, it has been shown that partial or complete loss of function alleles of IL-12/IFN-γ axis genes associated with diseases development.[13,14,34] In the present study, IFN2109G allele was significantly associated with increased susceptibility to TB. However, we found no significant association between IFNR1611 A/G SNP and TB patients. Previously, Mirsaeidi et al., also did not find any significant association between IFNR1395 SNP and susceptibility to TB among Iranian studied cases.[35] Also few studies declines the correlation of IFNGR1 polymorphism with M. tuberculosis, instead they proposed the correlation of IFNGR1 polymorphism with avirulent or M. bovis BCG infection.[25,36] These observations may outline the alternative pathways for enhancing host immune response against M. tuberculosis.
Conclusions
Our findings showed that the polymorphisms in TNF-α promoter gene are likely associated with increased susceptibility to TB in Iranian patients. But, no significant association was found between frequencies of SNPs in host and genotyping of M. tuberculosis. However, further studies with multiple genes polymorphisms would be necessary to elucidate the exact role of M. tuberculosis genotyping.
Acknowledgements
We thank all the TB patients and their families who have patiently helped us to complete the required information. The project was founded by MRC/NRITLD/WHO grant no, 0219-28-2010. There is no competing interest.
References

































[TOP]