Serena Vita1, Emanuela Giombini1, Patrizia De Marco1, Martina Rueca1, Cesare Ernesto Maria Gruber1, Alessia Beccacece1, Laura Scorzolini1, Valentina Mazzotta1, Carmen Pinnetti1, Priscilla Caputi1, Daniele Focosi2, Enrico Girardi1, Andrea Antinori1, Fabrizio Maggi1, Alessandra D’Abramo1# and Emanuele Nicastri1 and Spallanzani COVID-19 case investigation team*.
1 National Institute for Infectious Diseases “Lazzaro Spallanzani” IRCCS- Rome Italy.
2 North-Western Tuscany Blood Bank, Pisa University Hospital, Pisa, Italy.
Correspondence to:
Alessandra D’Abramo. National Institute for Infectious Diseases
“Lazzaro Spallanzani” IRCCS via Portuense 292, 00149 Rome Italy.
E-mail:
alessandra.dabramo@inmi.it
Published: May 01, 2024
Received: February 19, 2024
Accepted: April 18, 2024
Mediterr J Hematol Infect Dis 2024, 16(1): e2024043 DOI
10.4084/MJHID.2024.043
This is an Open Access article distributed
under the terms of the Creative Commons Attribution License
(https://creativecommons.org/licenses/by-nc/4.0),
which permits unrestricted use, distribution, and reproduction in any
medium, provided the original work is properly cited.
|
To the editor
Immunocompromised
(IC) patients are at higher risk for persistent and/or severe
SARS-CoV-2 infection caused by different viral variants, with a high
case-fatality ratio.[1,2] The first persistent
SARS-CoV-2 infection (5 months) was reported in 2020 in an IC patient
with a long persistence of SARS-CoV-2,[3] immediately followed by further reports.[2,4]
Indeed, the impairment of the immune system changes the natural history
of COVID-19. However, no consensus exists on clinical management of IC
COVID-19 patients.[5] Several reports emphasize the
clinical relevance of a combination therapy between small-molecule
antivirals (AV) and anti-spike monoclonal antibodies (MoAbs) both in
early and prolonged COVID-19 clinical management.[6,7]
In 2022, tixagevimab/cilgavimab (T/C) MoAb fixed combination was
introduced as early therapy for outpatient with COVID-19.[8]
We describe here a single-center case series of 22 IC COVID-19 in
patients with hematological disorders (HD) treated with a combined
therapy based on tixagevimab/cilgavimab (T/C) plus small-molecule
antivirals (AV), between April 1, 2022, and November 30, 2022.
The
viral genomic evolution was assessed by sequencing the whole SARS-CoV-2
genome in a subgroup of patients (pts). Pts were consecutively admitted for
COVID-19 to the Lazzaro Spallanzani National Institute for Infectious
Diseases, Rome, Italy (INMI). Demographic characteristics, medical
history, clinical presentation, treatment, adverse drug reactions, and
clinical outcome (survival/death) during follow-up were collected from
patient clinical records. Real-time reverse transcription polymerase
chain reaction (RT-PCR) on nasopharyngeal swab (NPS) samples was
performed according to the laboratory workflow using Alinity m
SARS-CoV-2 Assay (Abbott, Chicago, Illinois, United States) targeting
RdRp and N genes. When possible, molecular characterization of the
SARS-CoV-2 virus was performed using whole genome sequencing (WGS) at
diagnosis and during follow-up.[9] Whole Genome
sequencing (WGS) was carried out on an Ion Torrent Gene Studio S5
platform using Ion AmpliSeq SARS-CoV-2 in-sight research assay
following the manufacturer’s instructions (ThermoFisher Scientific,
Waltham, MA, USA). The whole genome reconstruction was performed using
ESCA software.[10] All the mutations were identified
with respect to the reference suggested by NCBI Wuhan-Hu-1
(NC_045512.2). A phylogenetic tree was built using 16 Italian
SARS-CoV-2 sequences that were selected among those available on the
GISAID platform with a collection date closer to that of the INMI
patients and clustered using cd-hit with 99% identity.[11]
The transition model (TIM+I+F+G) was identified as the best-fitting
nucleotide substitution model, and a phylogenetic tree was constructed
with 5,000 bootstrap replications using the IQ-Tree program.[12]
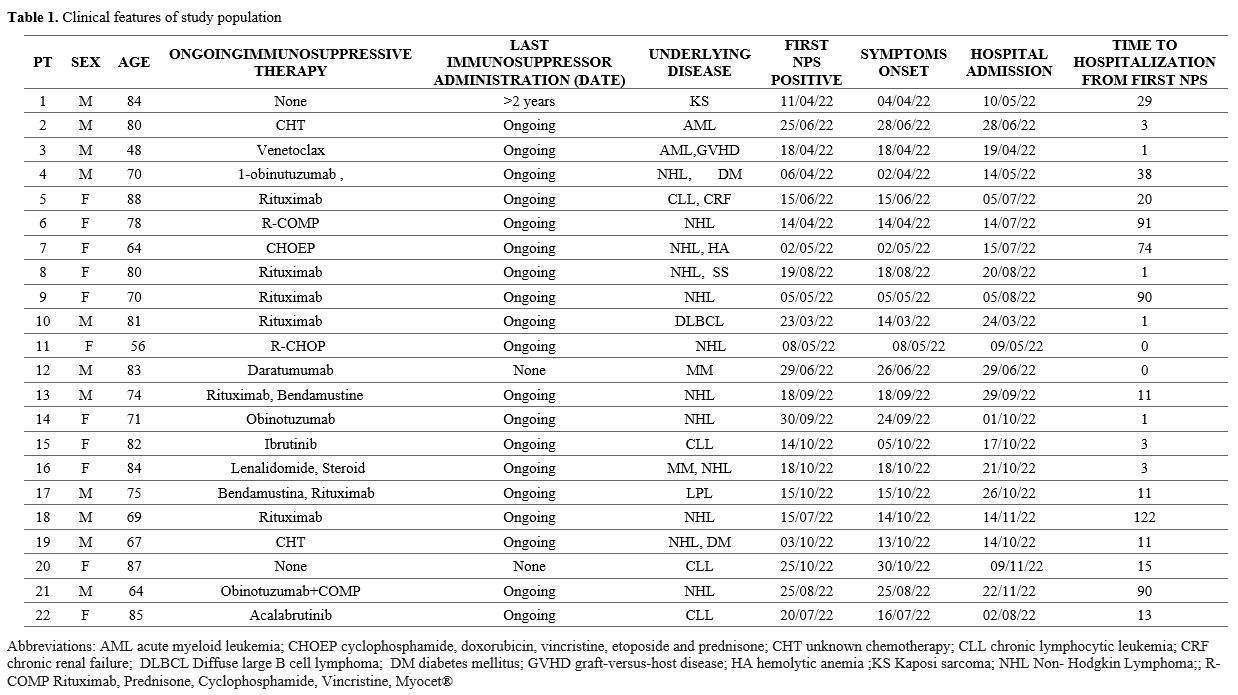
Table 1 shows
the characteristics of the study population. All patients were fully
vaccinated against COVID-19 with at least 3 doses, 11 (50%) of them
males, with a median age of 78 years old (IQR 69-83) (Table 1).
 |
- Table
1. Clinical features of study population
|
Twenty
patients were under active chemotherapy. They were admitted with a
median of 11 days (IQR 1-33) after the first NPS positive for
SARS-CoV-2. The study population had a median total lymphocyte count of
910/µl (IQR 520-1547), and 15 out of 22 (68%) had
hypogammaglobulinemia. All patients had pneumonia, but only 14 of them
required respiratory support. Seven patients had severe COVID-19 (WHO
COVID-19 ordinary scale 5), and 15 patients had moderate/mild COVID-19
(6 patients with a score of 4 and 9 patients with a score of 3).
Steroid therapy (oral or intravenous 6 mg dexamethasone daily) was
started in 14 patients with respiratory failure. At the admission, NPS
for SARS-CoV-2 was positive with a median cycle threshold (Ct) of 20
(IQR 16-24). All patients were treated with a first combination regimen
of MoAbs (T/C in 17 cases, sotrovimab in 3 cases, and
casirivimab/imdevimab in 2 cases) plus a 5-day course of intravenous
remdesivir (200 mg on day one followed by 100 mg on day 2-5). Eleven
out of 22 (50%) patients with an NPS<35 Ct required a second course
of antivirals (remdesivir in 2 cases and oral nirmatrelvir/ritonavir in
9 cases, 300mg/100 mg twice daily for 5 days) associated with T/C in
the five subjects initially treated with different MoAbs. Two patients
who, after 2 courses of antivirals and T/C, still had an NPS<35 Ct
received at least 2 doses of COVID-19 convalescent plasma (CCP) with
> 1:160 SARS-CoV-2 neutralizing antibody titer. Four patients died
(all with positive NPS PCR at the last available time point, i.e., at
days 103, 115, 43, and 41, respectively, since the first positive NPS)
(see Table 2). In particular:
•
Patient #3 died of gastrointestinal severe graft-versus-host diseases
(GvHD) at month 2 after hematopoietic stem cell transplantation for
acute myeloid leukemia.
• Patient #9 died of recurrent Clostridioides difficile infection during a relapse of NHL.
• Patient #19 died from a relapse of NHL.
•
Patient #22 died of respiratory failure and pneumonia sustained by
Aspergillus spp. and Stenotrophomonas maltophilia.
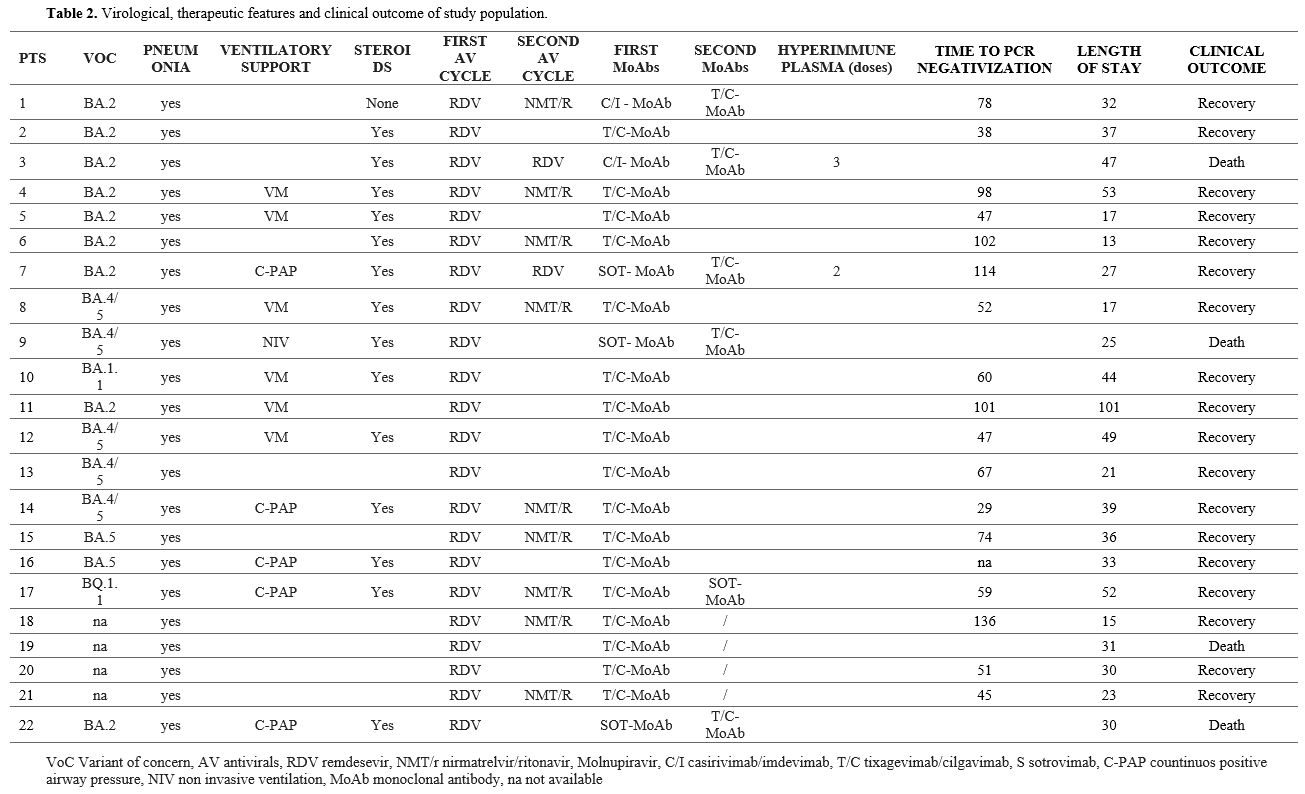
In the
remaining 18 patients, the SARS-COV-2 NPS PCR was negative at a median
of 59 (IQR 47-93) days since the first evidence of SARS-CoV-2 infection
(Table 2) and 47 days (IQR 28-51) after starting the treatment. The
median duration of hospital stay was 32 days (IQR 24-41).
 |
- Table 2. Virological, therapeutic features and clinical outcome of study population.
|
Spike-gene
sequencing was possible in 18 out of 22 patients, and identified a
BA.2* VoC in 9, a BA.4/5* VoC in 7, a BA.1.1* in 1, and a BQ.1.1* VoC
in 1.
The whole SARS-CoV-2 genome was sequenced in 4 out of 22
BA.2 patients (Patient#1, #3, #4, and #7). A deeper analysis was
conducted on the Spike glycoprotein. No recurrent amino acid mutations
in the 21 sequenced patients were found. In baseline sequences, no
mutations that were not lineage-related were found in patients #3 and
#4 (Table 3), while V445A
mutation in patient #1 and E340Q, R683W, and G798S mutations in patient
#7 were found. Patients #1 and #4 exhibited 3 and 1 additional Spike
mutations at the available second timepoint (T1), compared to the
baseline sequences. In particular, T1 patient #1 sequence showed a
deletion in position S: A243-L244. Finally, the phylogenetic tree
showed that whole genome sequences collected at baseline clustered with
a significative bootstrap with sequences collected after days 22 and 80
for patients #1 and #4, respectively, while the baseline sequence of
patient #3 was interspersed between other BA.2 sequences currently
circulating in Italy (Figure 1).
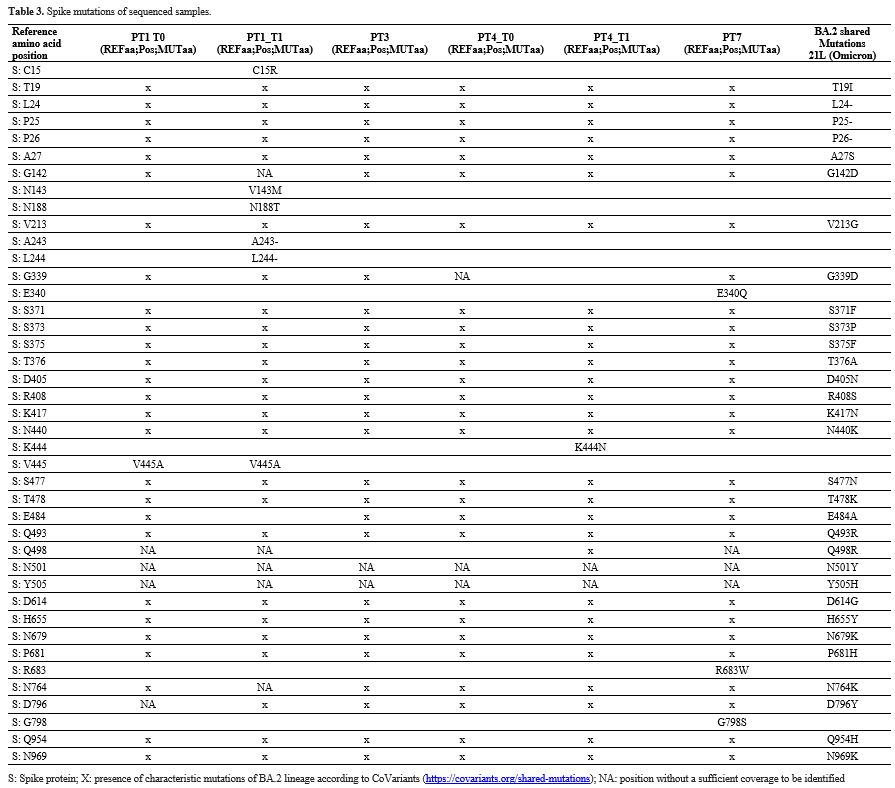 |
Table 3. Spike mutations of sequenced samples. |
 |
Figure 1. Phylogenetic analysis on whole genome sequences
|
In
the context of SARS-CoV-2 infection, IC patients face heightened
vulnerability. Although they have been underrepresented in previous
randomized clinical trials, they are likely overrepresented among
currently hospitalized patients with severe and/or persistent symptoms
associated with SARS-CoV-2 infection.[5,13]
Nevertheless, there is no evidence-based approach for managing these
patients. Several recent studies support the use of MoAb and AV
combination therapy in IC inpatients and outpatients or, for
inpatients, prolonged antiviral therapy.[7,14-18]
At admission, the cohort had a median of 11 days since the first
SARS-CoV-2 positive NPS, with a median Ct value of 20, suggesting a
persistently high viral replication. Notably, Ct-values, a measure of
viral burden, between 17 and 32 represent an amount of virus that is
likely to be replicative competent.[19] Seventy-one
percent of patients had a BA.2* VoC that retains in-vitro
susceptibility to cilgavimab; T/C has reduced efficacy against BA.5*
VoC, although it was unclear at that time of use. All patients were
considered at high risk of clinical progression and underwent a full
course of remdesevir and MoAb combined therapy with an off-label 600 mg
tixagevimab/cilgavimab prescription with no reported adverse event.
Half of them achieved viral clearance after the first course of
treatment, whereas the remaining 11 patients necessitated a second AV
and MoAb combined course.
Additionally, two patients only
partially responder (NPS<35 Ct) after two full combined antiviral
regimens, received CCP, a major therapeutic option as a source of
exogenous specific antibodies against SARS-CoV-2 Spike glycoprotein:
one patient died, and one recovered. We considered the 35 Ct cut-off
value during therapy as a surrogate marker of successful viral
response. Lower Ct values are commonly related to active viral
replication and potential contagiousness.[19,20]
All
COVID-19 survival patients had a negative SARS-CoV-2 NPS PCR after
combined therapy, with a median time of 52 days since the first
positive NPS and of 38 days since hospitalization. The observed case
fatality rate in our cohort was 18%, which falls within the previously
reported range of 13.8% to 39%.[21] The four deceased
patients tested positive for NPS PCR at the time of death: in three
patients, the death was due to recurrence of the underlying HD, and in
one case, to complication of stem cell transplant.
The
literature poorly describes IC patients treated by T/C, and this MoAb
has provided new therapeutic opportunities apart from the already two
registered indications.[8] Lahouati describes the
treatment of a cohort of 223 IC patients, although patients with HD
represented 25%, and among them, 12% were treated with T/C,
corresponding to 7 pts.[22]
In our cohort, all
patients were fully vaccinated against SARS-CoV-2. Indeed, COVID-19
vaccination among IC persons has been found to be highly protective
against COVID–19–associated hospitalization, leading to fewer
hospitalized patients and deaths.[23] All surviving
patients were able to resume treatment for their underlying disease a
few weeks after SARS-CoV-2 viral clearance. Although the molecular
analysis was performed only in four patients, it showed that affected
viruses did not contain any recurrent mutation present in all samples.
This suggests that in the 4 sequenced patients, there was no specific
mutation pattern that could be associated with the reported long
shedding or clinical severity. Although the analysis of a second-time
point was possible in only two patients, the follow-up mutation profile
of patients #1 and #4 was consistent with the observations of Leung.[2]
Patient #4 had a lower number of new mutations than patient #1,
considering that the interval period between the two sampling was 80
and 22 days, respectively (Table 3).
The V445A variant of SARS-CoV-2 Spike was found in patient #1 at both
time points. This mutation is located within the ACE2 receptor-binding
domain (RBD; aa 438-506) and causes full resistance to imdevimab and
bebtelovimab[24] and partial resistance to but did not induce immune evasion to casirivimab.[25]
In the second sampling of patient #4, the additional S: K444N mutations
within the RBD were reported, which reduces neutralization by
bebtelovimab[26] and imdevimab. A S: E340Q baseline mutation was reported in patient #9, which causes resistance to sotrovimab.[27]
Our
case series showed that in IC patients, the use of AV combined with
passive immunotherapy (MoAbs or CCP) is safe and can be effective.
Indeed, AV blocks viral replication, while MoAbs or CCP directed to the
Spike protein can neutralize the ability of the virus to bind and fuse
with the target host cell, reduce cytokine storm intensity in COVID-19
patients, and alleviate symptoms.[28] Finally,
combined antiviral therapy can reduce or completely limit the emergence
of drug-resistant mutations during prolonged sequential antiviral
monotherapy and is superior to monotherapy in terms of viral clearance.[6,7,14,15,29]
The
study acknowledges limitations inherent to its retrospective,
single-center design and restricted sample size. Additionally, the
small cohort hinders the ability to analyze the impact of specific
variables like hematological disorder types or disease severity.
Furthermore, whole genome sequencing data, offering a more
comprehensive analysis of viral strains, was only available for a
subset of patients.
Despite being a small case series, this study
offers valuable insights into a critical gap: the underrepresentation
of immunocompromised patients with HD in COVID-19 clinical trials. The
findings suggest a potential link between active HD and higher
mortality in IC COVID patients, even with mild symptoms. This
underscores the importance of treating all IC COVID patients with HD
and the need for further research on standardized combination therapies
for this population.
Author Contributions
Conceptualization,
SV, EmG, AD'A; Data curation, PD, AB, PC; Funding acquisition, EN,
CEMG; Investigation, GM, LS, VM, CP; Experiments, EmG, MR, and CEMG;
Supervision, EN, DF and FM; Validation, EN, EG, FV; Writing-original
draft, SV, EG, AD'A and EN; Writing-review and editing, AA, LS, MR,
CEMG, FM, DF and EN. All authors contributed to the article and
approved the submitted version. All authors have read and agreed to the
published version of the manuscript.
Funding
This
work was supported by Line1 Ricerca Corrente “Studio dei patogeni ad
alto impatto sociale: emergent, da importazione, multiresistenti,
negletti” funded by Italian Ministry of Health, and 5 per Mille-
Progetto 5M-2020-23682104.
Institutional Review Board Statement
Since the retrospective nature of our data, ethical approval was not required.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available by request to the corresponding author.
Acknowledgments
Spallanzani
COVID-19 Case Investigation Team: Tommaso Ascoli Bartoli, Nazario
Bevilacqua, Angela Corpolongo, Ambrogio Curtolo, Francesca Faraglia,
Maria Letizia Giancola, Gaetano Maffongelli, Claudia Palazzolo, Andrea
Mariano, Silvia Rosati, Maria Virginia Tomassi.
References
- Singson JRC, Kirley PD, Pham H, Rothrock G,
Armistead I, Meek Jet al. Factors Associated with Severe Outcomes Among
Immunocompromised Adults Hospitalized for COVID-19 - COVID-NET, 10
States, March 2020-February 2022. MMWR Morb Mortal Wkly Rep. 2022 July
8;71(27):878-884. https://doi.org/10.15585/mmwr.mm7127a3 PMid:35797216 PMCid:PMC9290380
- Leung
WF, Chorlton S, Tyson J, Al-Rawahi GN, Jassem AN, Prystajecky N, et al.
COVID-19 in an immunocompromised host: persistent shedding of viable
SARS-CoV-2 and emergence of multiple mutations: a case report. Int J
Infect Dis. 2022 Jan;114:178-182. https://doi.org/10.1016/j.ijid.2021.10.045 PMid:34757008 PMCid:PMC8553657
- Choi
B, Choudhary MC, Regan J, Sparks JA, Padera RF, Qiu X, et al.
Persistence and Evolution of SARS-CoV-2 in an Immunocompromised Host. N
Engl J Med. 2020 December 3;383(23):2291-2293. https://doi.org/10.1056/NEJMc2031364 PMid:33176080 PMCid:PMC7673303
- D'Abramo
A, Vita S, Maffongelli G, Beccacece A, Agrati C, Cimini E, et al.
Clinical Management of Patients With B-Cell Depletion Agents to Treat
or Prevent Prolonged and Severe SARS-COV-2 Infection: Defining a
Treatment Pathway. Front Immunol. 2022 May 27;13:911339. https://doi.org/10.3389/fimmu.2022.911339 PMid:35711444 PMCid:PMC9196078
- D'Abramo
A, Vita S, Nicastri E. Correction: The unmet need for COVID-19
treatment in immunocompromised patients. BMC Infect Dis. 2023 January
31;23(1):61. Erratum for: BMC Infect Dis. 2022 December 12;22(1):930. https://doi.org/10.1186/s12879-023-08034-0 PMid:36721126 PMCid:PMC9888749
- Orth
HM, Flasshove, C, Berger M, Hattenhuauer T, Biederbick KD, Mispelbaum R
et al. Early combination therapy of COVID-19 in high-risk patients.
Infection. 2023; https://doi.org/10.1007/s15010-023-02125-5 PMCid:PMC10955030
- D'Abramo
A, Vita S, Beccacece A, et al. B-cell-depleted patients with persistent
SARS-CoV-2 infection: combination therapy or monotherapy? A real-world
experience. Front Med (Lausanne). 2024 February 29;11:1344267. https://doi.org/10.3389/fmed.2024.1344267 PMid:38487021 PMCid:PMC10937561
- Vita
S, Rosati S, Ascoli Bartoli T, Beccacece A, D'Abramo A, Mariano A, et
al. Monoclonal Antibodies for Pre- and Postexposure Prophylaxis of
COVID-19: Review of the Literature. Pathogens. 2022 Aug 5;11(8):882. https://doi.org/10.3390/pathogens11080882 PMid:36015003 PMCid:PMC9412407
- Berno
G, Fabeni L, Matusali G, Gruber CEM, Rueca M, Giombini E, et al.
SARS-CoV-2 Variants Identification: Overview of Molecular Existing
Methods. Pathogens. 2022 Sep 17;11(9):1058. https://doi.org/10.3390/pathogens11091058 PMid:36145490 PMCid:PMC9504725
- Rueca
M, Giombini E, Messina F, Bartolini B, Di Caro A, Capobianchi MR, et
al. The Easy-to-Use SARS-CoV-2 Assem-bler for Genome Sequencing:
Development Study. JMIR Bioinform Biotech. 2022 Mar 14;3(1):e31536. https://doi.org/10.2196/31536 PMid:35309411 PMCid:PMC8924907
- Fu
L, NiuB, Zhu Z, Wu S, & Li W. CD-HIT: accelerated for clustering
the next-generation sequencing data. In Bioinformatics. 2012; (Vol. 28,
Issue 23, pp. 3150-3152). Oxford University Press (OUP). https://doi.org/10.1093/bioinformatics/bts565 PMid:23060610 PMCid:PMC3516142
- Nguyen
LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective
stochastic algorithm for estimating maximum-likelihood phylogenies. Mol
Biol Evol. 2015 Jan;32(1):268-74. https://doi.org/10.1093/molbev/msu300 PMid:25371430 PMCid:PMC4271533
- Trøseid
M, Hentzien M, Ader F, Cardoso SW, Arribas JR, Molina JM, et al..
Immunocompromised patients have been neglected in COVID-19 trials: a
call for action. Clin Microbiol Infect. 2022; 28:1182-3. https://doi.org/10.1016/j.cmi.2022.05.005 PMid:35623577 PMCid:PMC9130310
- Hirai
J, Mori N, Sakanashi D, Ohashi W, Shibata Y, Asai N, et al. Real-World
Experience of the Comparative Effectiveness and Safety of Combination
Therapy with Remdesivir and Monoclonal Antibodies versus Remdesivir
Alone for Patients with Mild-to-Moderate COVID-19 and
Immunosuppression: A Retrospective Single-Center Study in Aichi, Japan
[Internet]. Viruses. 2023; MDPI AG; 2023. p. 1952. https://doi.org/10.3390/v15091952 PMid:37766358 PMCid:PMC10538070
- Calderón-Parra
J, Gutiérrez-Villanueva A, Ronda-Roca G, Jimenez MLM, de la Torre H,
Ródenas-Baquero M, et al. Efficacy and safety of antiviral plus
anti-spike monoclonal antibody combination therapy vs. monotherapy for
high-risk immunocompromised patients with mild-to-moderate SARS-CoV2
infection during the Omicron era: A prospective cohort study. Int J
Antimicrob Agents. 2024 Mar;63(3):107095. https://doi.org/10.1016/j.ijantimicag.2024.107095 PMid:38244814
- Mikulska
M, Sepulcri C, Dentone C, Magne F, Balletto E, Baldi F, et al. Triple
Combination Therapy With 2 Antivirals and Monoclonal Antibodies for
Persistent or Relapsed Severe Acute Respiratory Syndrome Coronavirus 2
Infection in Immunocompromised Patients [Internet]. Clinical Infectious
Diseases. 2023; p. 280-6. https://doi.org/10.1093/cid/ciad181 PMid:36976301
- Brosh-Nissimov
T, Ma'aravi N, Leshin-Carmel D, Edel Y, Ben Barouch S, Segman Y, et al.
Combination treatment of persistent COVID-19 in immunocompromised
patients with remdesivir, nirmaltrevir/ritonavir and
tixegavimab/cilgavimab. J Microbiol Immunol Infect. 2024;57(1):189-194.
https://doi.org/10.1016/j.jmii.2023.09.004 PMid:37805361
- Vita
S, D'Abramo A, Coppola A, Farroni C, Iori A P, Faraglia F,et al.
Combined antiviral therapy as effective and feasible option in
allogenic hematopoietic stem cell transplantation during SARS-COV-2
infection: a case report. Frontiers in Oncology. 2024;14.22 https://doi.org/10.3389/fonc.2024.1290614 PMid:38414746 PMCid:PMC10896944
- Wölfel
R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, et al.
Virological assessment of hospitalized patients with COVID-2019.
Nature, 2020 May;581(7809):465-469. Epub 2020 April 1. Erratum in:
Nature. 2020 ;588(7839):E35. https://doi.org/10.1038/s41586-020-2196-x PMid:32235945
- Singanayagam
A, Patel M, Charlett A, Bernal JL, Saliba V, Ellis J et al. Duration of
infectiousness and correlation with RT-PCR cycle threshold values in
cases of COVID-19, England, January to May 2020. Euro Surveill.
2020;25. https://doi.org/10.2807/1560-7917.ES.2020.25.32.2001483 PMid:32794447 PMCid:PMC7427302
- Pagano
L, Salmanton-García J, Marchesi F, Busca A, Corradini P, Hoenigl M, et
al. COVID-19 infection in adult patients with hematological
malignancies: a European Hematology Association Survey (EPICOVIDEHA). J
Hematol Oncol. 2021;14(1):168. https://doi.org/10.1186/s13045-021-01177-0 PMid:34649563 PMCid:PMC8515781
- Lahouati
M, Cazanave C, Labadie A, Gohier P, Guirlé L, Desclaux A, et al;
Bordeaux COVID-19 Treatment Group. Outcomes of targeted treatment in
immunocompromised patients with asymptomatic or mild COVID-19: a
retrospective study. Sci Rep. 2023 Sep 16;13(1):15357. https://doi.org/10.1038/s41598-023-42727-5 PMid:37717101 PMCid:PMC10505186
- Farroni
C, Aiello A, Picchianti-Diamanti A, Laganà B, Petruccioli E, Agrati C,
et al. Booster dose of SARS-CoV-2 messenger RNA vaccines strengthens
the specific immune response of patients with rheumatoid arthritis: A
prospective multicenter longitudinal study. Int J Infect Dis.
2022;125:195-208. https://doi.org/10.1016/j.ijid.2022.10.035 PMid:36328289 PMCid:PMC9622025
- Focosi
D, Maggi F, Franchini M, McConnell S, Casadevall A. Analysis of Immune
Escape Variants from Anti-body-Based Therapeutics against COVID-19: A
Systematic Review. Int J Mol Sci. 2021 Dec 21;23(1):29. https://doi.org/10.3390/ijms23010029 PMid:35008446 PMCid:PMC8744556
- Cox
M, Peacock TP, Harvey WT, Hughes J, Wright DW; COVID-19 Genomics UK
(COG-UK) Consortium et al. SARS-CoV-2 variant evasion of monoclonal
antibodies based on in vitro studies. Nat Rev Microbiol.
2023;21(2):112-124. https://doi.org/10.1038/s41579-022-00809-7 PMid:36307535 PMCid:PMC9616429
- https://www.fda.gov/media/156152/download last accessed February 9, 2024
- Andrés
C, González-Sánchez A, Jiménez M, Márquez-Algaba E, Piñana M,
Fernández-Naval C, et al. Emergence of Del-ta and Omicron variants
carrying resistance-associated mutations in immunocompromised patients
undergoing sotrovimab treatment with long-term viral excretion. Clin
Microbiol Infect. 2022; S1198-743X(22)00458-X.
- Taylor
PC, Adams AC, Hufford MM, de la Torre I, Winthrop K, Gottlieb RL.
Neutralizing monoclonal antibodies for treatment of COVID-19. Nat Rev
Immunol. 2021;21(6):382-393. https://doi.org/10.1038/s41577-021-00542-x PMid:33875867 PMCid:PMC8054133
- De
Forni D, Poddesu B, Cugia G, Chafouleas J, Lisziewicz J, Lori F.
Synergistic drug combinations designed to fully suppress SARS-CoV-2 in
the lung of COVID-19 patients. PLoS One. 2022 Nov 10;17(11):e0276751. https://doi.org/10.1371/journal.pone.0276751 PMid:36355808 PMCid:PMC9648746



