The use of metagenomics could identify the presence of pathogenic species and resistance genes in the urinary tract of at-risk individuals and guide subsequent antibiotic therapy, reducing unnecessary exposure to broad-spectrum antibiotics.[4] Although “snapshots” may contribute to elucidating factors that perturb the microbiota and what impact these changes have on the development of disease, the evaluation of longitudinal data may be useful in establishing the significance of these changes.
In this context, several investigations have failed to address the association between urinary microbiota and long-term patient outcomes.[5] Spinal dysraphism (also known as spina bifida) causes neurological deficits, including neurogenic bladder, resulting in detrusor sphincter dyssynergia. This condition represents a predisposing factor for urinary tract infections (UTIs).[6,7]
In this work, we routinely followed up a pediatric cohort with spinal dysraphism for 3 years after microbiota profiling to establish any correlation between this and the development of subsequent UTIs.
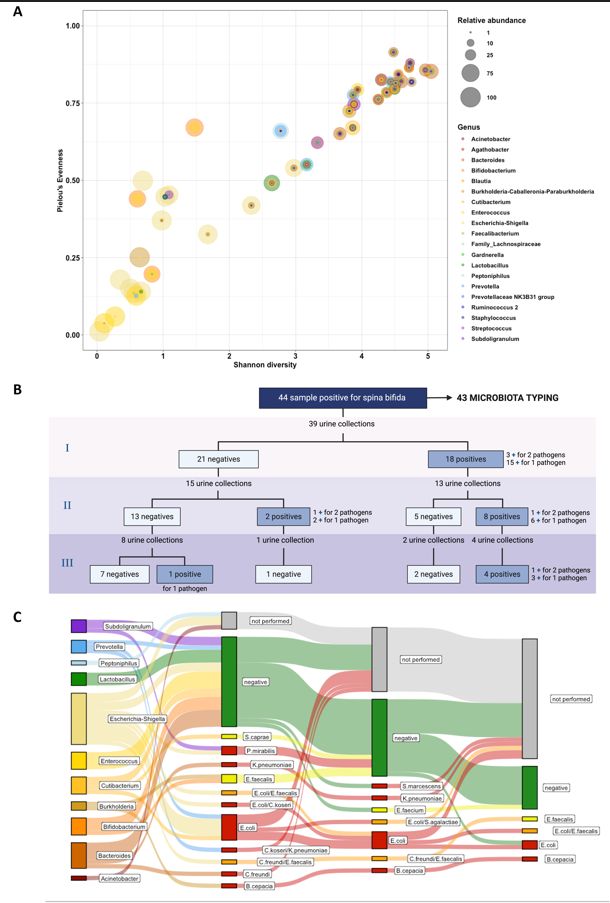
We studied microbiota in a cohort that included individuals who were not taking antibiotics or probiotics and did not have a UTI at the time of recruitment.[8] The relative abundance of neuropathological species in samples from patients with spina bifida increased over time, while patients routinely performing clean intermittent catheterization (CIC) had a predominance of skin organisms. 16s sequencing raw data obtained from 43/44 specimens were analyzed at the genus level, selecting the top 20 genera identified in the dataset. Relative abundances were then matched with Shannon diversity and Pielou’s Evenness (Figure 1A). Alpha diversity metrics, as expected, showed a positive Pearson correlation (p<0.001, Rho=0.924), suggesting that with the decrease in species number, one or more microbes prevailed in the community. Low values of both alpha diversity metrics were associated with a higher relative abundance of genera manifestly recognized as uropathogens.
We followed up on these patients for 3 years and carried out a relationship between urine culture results and the most abundant genera. 39/44 individuals had a urine culture examination. 18 samples were positive at the first collection (median = 12 months); 10/28 positive samples at the second time point (median = 11 months), 2 of whom belonged to previous negative samples; 5/15 positive samples at the third urine collection (median = 11 months), one of whom belonged to previous negative patients (Figure 1B).
Interestingly, 12/43 (28%) samples showed E. coli-Shigella as the most abundant genus, 8 of whom (67%) were associated with culture-positive samples at the first follow-up (6 samples for gram-negative bacteria and 2 samples for both gram-negative and gram-positive bacteria). While the Burkholderia genus was always associated with a positive urine sample, Bacteroides, Cutibacterium, Prevotella, and Subdoligranulum genera were only partially associated with a positive sample at the first follow-up. Interestingly, samples dominated by Enterococcus, Lactobacillus, and Bifidobacterium genera appeared mostly associated with negative urine samples (Figure 1C). These findings suggested that the prevalence of specific genera may have a protective activity against UTIs, leading to even reevaluating the role of some bacteria. Enterococcus and Lactobacillus are two of the most renowned genera that produce antimicrobial peptides (such as bacteriocins).[9] Of note, 15/18 of urine positive at the first follow-up belonged to patients managed with CIC, highlighting this as a major risk factor for the development of UTI. This result is opposite to what was shown by Kaye et al., suggesting a further deep clinical investigation in these patients.[6]
Although it is plausible that the small sample size might have influenced our results (including the sample drop-out during follow-up), our findings prompt 16s analysis as a promising candidate to investigate urinary tract infections in special at-risk populations as patients with neurogenic bladder, vesicoureteral reflux, renal abnormalities or a history of recurrent UTIs. While urine culture remains the gold standard for the diagnosis of urinary tract infections, the characterization of bladder microbiota may represent an innovative and additional diagnostic tool for the diagnosis of UTIs in vulnerable populations.[10,11]
On the other hand, further longitudinal studies will be needed to harmonize microbiota characterization with clinical outcomes, avoid the use of inappropriate antibiotic prophylaxis, and opportunely set conventional and non-conventional antimicrobial interventions.
Ethics approval and consent to participate
The study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of Policlinico Universitario “A. Gemelli” IRCCS for studies involving humans (ID: 4279). Written informed consent was obtained from each subject involved in the study.Funding
MS acknowledges EU funding for the MUR PNRR Extended Partnership initiative on Emerging Infectious Diseases (Project number PE00000007, INF-ACT).Authors’ contributions
This study was designed by F.D.M and C.R. Data collection and analysis by F.D.M. and G.S. Figure preparation was conducted by F.D.M. and G.S. F.D.M., G.S. and D.M.B. wrote the initial draft. M.S. and C.R. revised the initial draft. All authors participate in the discussion and interpretation of the results. All authors have read and agreed to the published version of the manuscript.Acknowledgments
We acknowledge the contribution of Microbiota analysis and Microbial WGS of the Fondazione Policlinico Universitario “A. Gemelli” IRCCS for sample processing and analysis.References
- Porcari S, Mullish BH,
Asnicar F, Ng SC, Zhao L,
Hansen R, O'Toole PW, Raes J, Hold G, Putignani L et al: International
consensus statement on microbiome testing in clinical practice. Lancet
Gastroenterol Hepatol 2025, 10(2):154-167. https://doi.org/10.1016/S2468-1253(24)00311-X
PMid:39647502
- Murray
BO, Flores C, Williams C, Flusberg DA, Marr EE, Kwiatkowska KM, Charest
JL, Isenberg BC, Rohn JL: Recurrent Urinary Tract Infection: A Mystery
in Search of Better Model Systems. Front Cell Infect Microbiol 2021,
11:691210. https://doi.org/10.3389/fcimb.2021.691210
PMid:34123879 PMCid:PMC8188986
- Perez-Carrasco
V, Soriano-Lerma A, Soriano M, Gutierrez-Fernandez J, Garcia-Salcedo
JA: Urinary Microbiome: Yin and Yang of the Urinary Tract. Front Cell
Infect Microbiol 2021, 11:617002. https://doi.org/10.3389/fcimb.2021.617002
PMid:34084752 PMCid:PMC8167034
- Kucherov
V, Russell T, Smith J, Zimmermann S, Johnston EK, Rana MS, Hill E, Ho
CP, Pohl HG, Varda BK: Antibiotic Overtreatment of Presumed Urinary
Tract Infection Among Children with Spina Bifida. J Pediatr 2024,
272:114092. https://doi.org/10.1016/j.jpeds.2024.114092
PMid:38734134
- Kawalec
A, Zwolinska D: Emerging Role of Microbiome in the Prevention of
Urinary Tract Infections in Children. Int J Mol Sci 2022, 23(2). https://doi.org/10.3390/ijms23020870
PMid:35055056 PMCid:PMC8775962
- Kaye
IY, Payan M, Vemulakonda VM: Association between clean intermittent
catheterization and urinary tract infection in infants and toddlers
with spina bifida. J Pediatr Urol 2016, 12(5):284 e281-284 e286. https://doi.org/10.1016/j.jpurol.2016.02.010
PMid:27118581
- Madden-Fuentes
RJ, McNamara ER, Lloyd JC, Wiener JS, Routh JC, Seed PC, Ross SS:
Variation in definitions of urinary tract infections in spina bifida
patients: a systematic review. Pediatrics 2013, 132(1):132-139. https://doi.org/10.1542/peds.2013-0557
PMid:23796735
- De
Maio F, Grotti G, Mariani F, Buonsenso D, Santarelli G, Bianco DM,
Posteraro B, Sanguinetti M, Rendeli C: Profiling the Urobiota in a
Pediatric Population with Neurogenic Bladder Secondary to Spinal
Dysraphism. Int J Mol Sci 2023, 24(9). https://doi.org/10.3390/ijms24098261
PMid:37175968 PMCid:PMC10178886
- Ben
Braiek O, Smaoui S: Enterococci: Between Emerging Pathogens and
Potential Probiotics. Biomed Res Int 2019, 2019:5938210. https://doi.org/10.1155/2019/5938210
PMid:31240218 PMCid:PMC6556247
- Sujith
S, Solomon AP, Rayappan JBB: Comprehensive insights into UTIs: from
pathophysiology to precision diagnosis and management. Front Cell
Infect Microbiol 2024, 14:1402941. https://doi.org/10.3389/fcimb.2024.1402941
PMid:39380727 PMCid:PMC11458535
- Fernandes
A, Jobby R: Bacteriocins from lactic acid bacteria and their potential
clinical applications. Appl Biochem Biotechnol 2022, 194(10):4377-4399.
https://doi.org/10.1007/s12010-022-03870-3
PMid:35290605