The association between P. vivax susceptibility and Duffy blood group status stemmed from observations that this malaria species was largely absent from West Africa, where over 95% of the population is Duffy-negative due to inheritance of the DARC-null FY*B null allele.[1,2] In contrast, high prevalence of Duffy-positive phenotypes persists across most regions of the Indian subcontinent, suggesting widespread susceptibility to infection by PvDBP-dependent P. vivax strains.[3] While the conventional paradigm held that Duffy-negative individuals were refractory to P. vivax blood-stage infection, sporadic case reports over the past decade in Duffy-negative populations from Botswana, Cameroon, Ethiopia, Equatorial Guinea, Mali, Mauritania, Senegal, Sudan, and Uganda challenged this biological rule.[4-12]
The first well-documented instances of Duffy-negative individuals harboring P. vivax infections emerged from the Brazilian Amazon region.[13,14] With an eye toward malaria elimination, increased prevalence surveillance soon revealed that over 6.4% of P. vivax cases in rural Amazonia involved Duffy-negative individuals.[15] Whole genome sequencing of P. vivax isolates from enigmatic infections subsequently uncovered molecular evidence for evolutionary selective sweeps in the pvdbp gene encoding the critical cytoadherence ligand.[16] A subset of Brazilian P. vivax parasites appeared to harbor PvDBP variants capable of mediating DARC-independent erythrocyte invasion, likely through the acquisition of mutations that expand receptor tropism.
Soon after, case reports of P. vivax infection of Duffy-negative individuals surfaced in Madagascar, Ethiopia, Equatorial Guinea, and other parts of sub-Saharan Africa traditionally deemed refractory to this malaria species.[3,17] Genomic analysis of African P. vivax isolates confirmed the evolution of PvDBP diversity distinct, with the emergence of novel duplications in pvdbp encoding non-canonical peptide signatures associated with DARC-independent invasion phenotypes.[16] Importantly, these alternative PvDBP variants retained capacity for DARC-dependent cytoadherence as well, suggesting that P. vivax strains can maintain the ability to infect both Duffy-positive and Duffy-negative populations.
While PvDBP diversification liberating P. vivax to infect Duffy-negative hosts has become increasingly apparent in parts of Africa and South America, the paradigm of DARC-dependent cytoadherence dominating P. vivax blood-stage biology has remained intact across most endemic regions of the Indian subcontinent. The Duffy-positive phenotype predominates in Indian populations, eliminating a key selective advantage for emergence of DARC-independent invasion pathways.[3]
Duffy-negative Indian individuals have been documented in hospital-based studies.[18,19] However, these cases had no evidence of P. vivax infection. Here, we describe Fynull infected with P. vivax from malaria-endemic regions across India.
Material and Methods
Ethics Information. Detailed information about the research was provided and explained to all the individuals who agreed to participate in the present study. All studies were performed in accordance with the recommendations put forth in the guide by the Institutional Ethics Committee Review Board and were approved by the Institutional Ethics Committee, Institutional Committee for Research on Human Subjects, National Institute of Immunohaematology (ICMR), Mumbai.Study site and patients. The present study collected 165 clinical blood samples from blood banks of different geographical regions of India (Mumbai (Mu, n=23) – Maharashtra state; Surat (Su. N=27) – Gujarat state; Manipur state (NE, n=90) and Mangalore (Ma, n=25) – Karnataka state). Peripheral blood was collected (3 cc. in EDTA) and stored at 4 ͦC till samples reached the testing centre. Samples found positives on a rapid diagnostic test (RDT) were sent to NIIH for molecular testing (Figure 1). Males and females aged over 18 years participated in the study. Information on the patient's ethnic background and other disease conditions, including diabetes and infectious disease (including malaria), was obtained by interviewing the individual. Healthy individuals' blood samples (n=200) were collected from respective regions as normal controls (Mumbai (Mu, n=30) – Maharashtra state; Surat state (Su. N=33) – Gujarat; Manipur state (NE, n=110), and Mangalore (Ma, n=27) - Karnataka state).
Serological testing. Duffy phenotyping was determined by haemagglutination assay using monoclonal antibody (Cat.: 3013-2; Immucor, Inc., Norcross, GA, USA) by the indirect antiglobulin test (IAT) in tubes and gel cards (Diamed SA, Morat, Switzerland) according to the manufacturer’s recommendations. Suitable controls were included during the serological phenotyping of RBCs.
Blood smear preparation and microscopic examination. Two thick and thin blood smear slides were prepared from fresh peripheral blood, air-dried, and preserved till Giemsa-staining, as described by WHO (2010).[20] The slides were sent to ICMR-NIIH for independent validation. All the smears were observed with oil immersion at a final magnification of x1000. The parasite density was calculated by counting the parasites and leucocytes.[21] The microscopic analyst was blinded by the results of RDT and molecular analysis. If no parasite was observed in oil immersion fields, the smear (sample) was considered negative. Parasite density was enumerated by calculating the number of parasites / 200 white blood cells (in a thick blood film).
DNA extraction from samples. DNA was extracted and purified from 200 μL of fresh blood using a QIAamp DNA Mini Kit (Qiagen, Venlo, Netherlands). After the purification step, the extracted DNA was eluted with 50 μL of elution buffer and quantified by Nanodrop-1000 (Thermo Fisher Scientific, Massachusetts, United States). The samples were diluted to attain a final concentration of 30 ng/µl.
Results
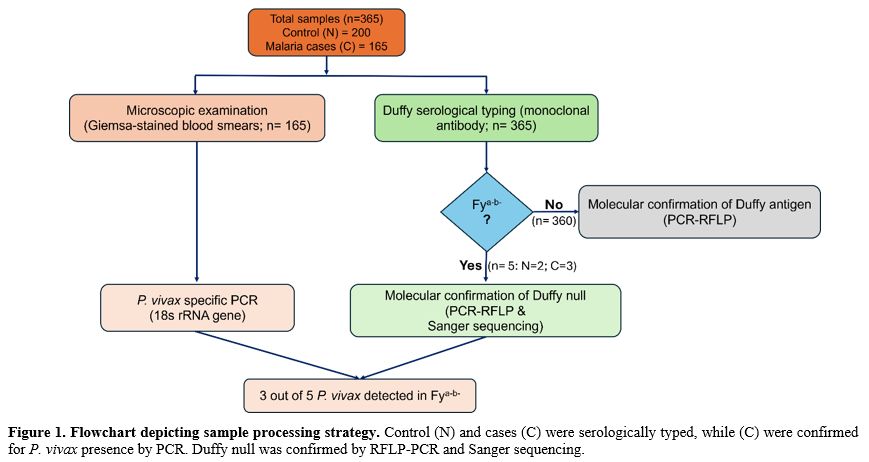
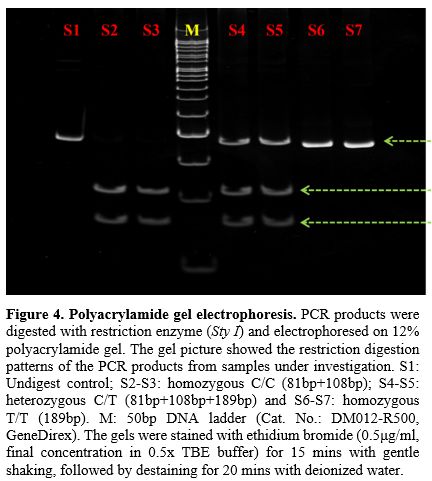
Serology. Three hundred sixty-five blood samples, which included 165 malaria-positive samples and 200 healthy donor samples, were tested using Duffy monoclonal antibody by standard tube technique. Of these, 360 samples showed normal Fya, Fyb, or Fya+b+ distribution, and 5 samples showed Duffy null (Fya-b-) phenotype (Figure 2). These Fy-negative samples, which showed no agglutination in the tube as well as under the microscopic field (Figure 3A), were also confirmed to be negative on gel cards (Figure 3B). This method provided initial screening for the Duffy antigen status (Fya-b-) of the subjects.PCR-RFLP - Genotyping. To confirm the Fy negative (Fya-b-) status observed by serological technique, we performed PCR-RFLP analysis on the 5 serologically Fy-negative samples. Duffy polymorphisms which included promoter GATA-1 box mutation c.–67T>C (FY* Null (FY*02N.01) mutation for detecting Fy null was identified by polymerase chain reaction-restriction fragment length polymorphism (PCR–RFLP) as described by Shaikh et al. (2024).[22] All five samples were analyzed by digestion using Sty I restriction enzyme, followed by running on a 12% polyacrylamide gel (Figure 4) and genotyped as Fya-b-. Control samples of confirmed genotypes were run alongside the samples under investigation. The RFLP pattern confirmed the absence of the Duffy antigen in all five individuals, corroborating the serological findings.
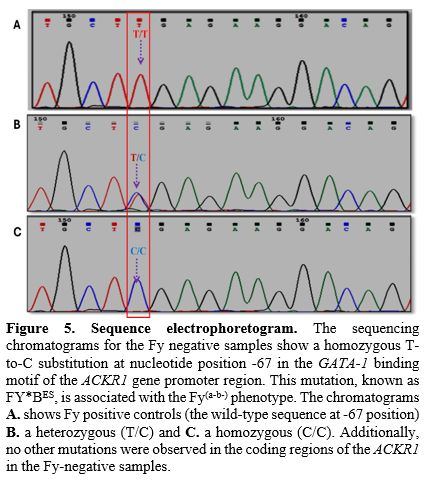
DNA Sequencing. We conducted Sanger sequencing on the relevant genomic region encompassing the GATA motif covering (T-67C) of the 5 Fy negative samples to further validate the Fy negative status and identify any potential mutations. Electrophoretogram analysis showed a homozygous T base at -67th position (upstream) of ACKR1 gene (Figure 5) in Duffy coding DNA sequence (CDS) when compared to the reference sequence (NM_002036.2). This indicates the absence of functional Duffy antigen expression in all 5 individuals, aligning with both the serological and PCR-RFLP findings.
Presence of P. vivax. Among the 5 Fy negative individuals, we detected P. vivax infection in 3 subjects (60%) using two complementary methods:
1. PCR: PCR-Specific PCR assays targeting the 18S rRNA gene of P. vivax (developed in-house) were performed on all malaria-positive samples along with appropriate controls (Figure 6). The results indicated the presence of P. vivax DNA in 3 out of the 5 Fy(a-b-) samples (60%).
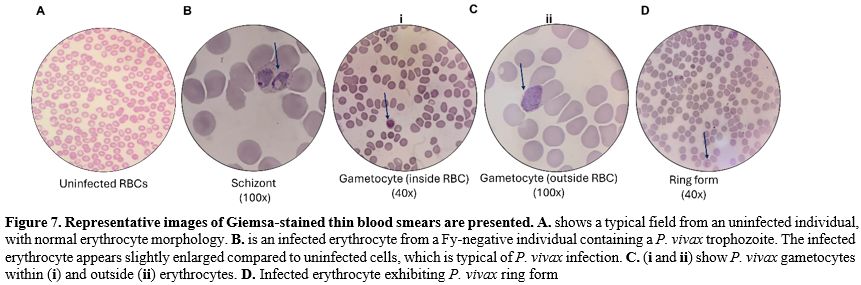
2. Microscopy: Thick and thin blood smears from all 5 Fy-negative individuals were prepared and stained with Giemsa for microscopic examination. Visual inspection of the stained slides confirmed the presence of P. vivax parasites in the same 3 Fy-negative individuals identified (Figure 7) as positive by PCR. Parasite stages observed included ring forms, trophozoites, and schizonts, with parasitemia ranging from 0.01% to 0.5%. These findings demonstrate the presence of P. vivax infections in Fy-negative individuals.
Discussion
The present study provides compelling evidence for P. vivax infection in Duffy-negative individuals, challenging the long-standing paradigm of Duffy antigen dependency for P. vivax invasion of human erythrocytes. Our findings, based on a combination of serological, microscopic, and molecular techniques, not only confirm the existence of Duffy-negative phenotypes but also demonstrate the presence of P. vivax parasites in these individuals.The three Duffy-negative individuals with confirmed P. vivax infection presented with distinct clinical features that provide valuable insights into parasite-host interactions in the absence of the conventional Duffy receptor pathway. They had fever lasting between 9 to 17 days with a characteristic tertian pattern - temperature rises every third day starting with a cold stage, followed by chills, rigor, fever, and finally diaphoresis. Body temperature peaked at 39°C during febrile episodes. All three patients exhibited splenomegaly, with palpable spleens extending one to three fingers below the costal margin, consistent with the immune response to malarial infection. Patient 1 (25-year-old male, Ahom tribe) presented with a 12-day history of fever with the most pronounced tertian periodicity. Patient 2 (32-year-old female, Ahom tribe) experienced a longer 17-day febrile illness with moderate anemia (hemoglobin 9.2 g/dL). Patient 3 (41-year-old male, Mangalore) had a 9-day history of fever with mild hepatomegaly in addition to splenomegaly. All three patients were treated with the standard artemisinin-based combination therapy (artesunate-lumefantrine) according to national guidelines. Following the fever resolution, they received a 7-day course of primaquine after G6PD deficiency was excluded, as per Government of India protocols for radical cure of P. vivax malaria. Clinical response to therapy was comparable to that typically observed in Duffy-positive individuals with P. vivax infection. The parasitemia levels in these Duffy-negative individuals ranged from 0.01% to 0.5%, which is within the typical range seen in P. vivax infections in the general population. This may suggest that despite using alternative invasion pathways, P. vivax parasites achieved comparable levels of blood-stage replication in Duffy-negative hosts.
Our serological screening identified 1.36% of the study population as Duffy-negative, consistent with previous reports from regions where this phenotype is present but not predominant.[3] The Duffy blood group system, determined by the ACKR1 gene, plays a crucial role in malaria susceptibility, particularly for P. vivax infections.[23] The Duffy-negative phenotype has been historically associated with protection against P. vivax malaria.[2]
The PCR-RFLP analysis and Sanger sequencing results confirmed the Duffy-negative status at the molecular level. The observed homozygous T-to-C substitution at nucleotide position -67 in the GATA-1 binding motif of the ACKR1 gene promoter region corresponds to the FY*BES allele, responsible for the Fy(a-b-) phenotype.[24] The presence of nucleotide T at -67th position in the promoter region (Figure 5) prevents binding of RBC-specific eFII factor and subsequent failure in the recruitment of RNA polymerase for the synthesis of mRNA, leading to the non-formation of ACKR1 antigen.[25]
The most striking finding of our study is the detection of P. vivax infections in 60% (3 out of 5) of the Duffy-negative individuals, consistently supported by both PCR analysis and microscopic examination of blood smears. The presence of various parasite stages in the infected Duffy-negative samples provides strong evidence for the parasite's ability to complete its erythrocytic cycle in these hosts.
This finding challenges the long-held belief that the Duffy antigen is an absolute requirement for P. vivax invasion of human erythrocytes.[26] While the Duffy antigen, specifically the Fy6 epitope, has been shown to interact with the P. vivax Duffy Binding Protein (PvDBP) during the invasion process;[27] our results suggest the existence of alternative invasion pathways or adaptations. The presence of 3 out of 5 Duffy-negative patients with P. vivax infection indicates that the resistance to Duffy-negative individuals for P. vivax infection is likely to be relative. In countries with intense P. vivax transmission, other modes of entry to red cells by merozoites may be used. Several recent studies have reported similar findings of P. vivax infections in Duffy-negative individuals from various geographic regions, including parts of Africa,[23] and South America.[17] Our study adds to this growing body of evidence and underscores the need to reevaluate our understanding of P. vivax host cell invasion mechanisms.
The ability of P. vivax to infect Duffy-negative individuals has profound implications for the parasite's biology and evolution. Several hypotheses have been proposed to explain this phenomenon: 1) Alternative receptors: P. vivax may be utilizing other receptors on the erythrocyte surface for invasion. Recent studies have identified additional P. vivax proteins, such as PvRBP2b, that can bind to receptors other than the Duffy antigen.[6] 2) Modifications to PvDBP: Genetic variations in the PvDBP gene may allow the protein to interact with other erythrocyte surface molecules or to bind to the Duffy antigen with higher affinity.[28] 3) Invasion of young erythrocytes: P. vivax may preferentially invade younger erythrocytes or reticulocytes that transiently express low levels of the Duffy antigen.[29]
The distribution of the Duffy-negative phenotype shows considerable variation across different ethnic groups globally, with the highest prevalence (>95%) in West African populations and lower rates in other regions.[3] In India, the Duffy-negative phenotype has been previously reported at low frequencies, primarily in specific ethnic groups and tribal populations.[18,19]
Our findings regarding the five Duffy-negative individuals identified in this study revealed interesting patterns related to ethnic background. Among the three Duffy-negative individuals infected with P. vivax, two belonged to the Ahom tribe (mongoloids) from Northeast India, while the third was from Udupi (Karnataka) in South India. The two Duffy-negative individuals in our control group (without P. vivax infection) were found to be from the Dedar community (Karnataka) and another from the Ahom tribe (mongoloids) in Northeast India (Figure 2). This distribution suggests possible genetic isolation and founder effects that may have contributed to the persistence of the Duffy-negative phenotype in these specific communities. The presence of Duffy-negative individuals in both the Ahom tribe of Northeast India and communities from Karnataka indicates that this phenotype, while rare, is distributed across geographically and ethnically distinct populations in India.
The identification of P. vivax infections in Duffy-negative individuals across different ethnic backgrounds (two from the Ahom tribe and one from Udupi) indicates that the phenomenon is not restricted to a single ethnic group in India. This observation is particularly significant as it suggests that the adaptation of P. vivax to infect Duffy-negative hosts may have occurred independently in different geographical regions of India or may represent the spread of adapted strains across diverse populations.
The ethnic distribution of Duffy-negative phenotypes in India likely reflects historical population movements, genetic drift, and possibly selective pressures related to malaria exposure. Certain tribal populations in India have remained relatively isolated and endogamous for generations, potentially preserving genetic variants at different frequencies compared to more admixed populations. The presence of the Duffy-negative phenotype across these diverse ethnic groups warrants further investigation into its evolutionary origins in the Indian subcontinent. Our findings highlight the importance of considering ethnic background in malaria epidemiological studies in India. The considerable genetic diversity across Indian populations, shaped by complex historical patterns of migration, admixture, and isolation, may influence susceptibility to different malaria species and strains. Large-scale population-based studies incorporating both Duffy phenotyping/genotyping and sensitive P. vivax detection methods across diverse ethnic groups would provide valuable insights into the true prevalence and distribution of Duffy-negative phenotypes and their relationship with P. vivax susceptibility in India. Furthermore, understanding the ethnic distribution of Duffy-negative phenotypes infected with P. vivax could have implications for targeted malaria control strategies, especially in regions with higher prevalence of specific ethnic groups known to carry the Duffy-negative trait. This knowledge could inform more effective surveillance, prevention, and treatment approaches tailored to the unique genetic landscape of different communities across India.
The adaptation of vivax infection in Duffy-negative individuals could have significant consequences for the global distribution and epidemiology of P. vivax malaria. Our findings, along with other recent reports, suggest that P. vivax may be more widespread in Africa and other regions with Duffy-negative populations than previously recognized.[30] This has several important consequences. The global burden of vivax malaria may be significantly underestimated.[16] P. vivax infections in Duffy-negative individuals may serve as a reservoir for ongoing transmission.[31] The adaptation of P. vivax to Duffy-negative hosts could facilitate the expansion of the parasite's geographical range.[32] Malaria elimination strategies may need to be reevaluated and adjusted to account for the possibility of transmission in Duffy-negative populations.[33]
The occurrence of P. vivax infections in Duffy-negative individuals also has important implications for malaria diagnosis and treatment. Healthcare providers should be aware of the possibility of P. vivax infections in Duffy-negative individuals.[34] management of P. vivax infections in Duffy-negative individuals may require special consideration.[35] Questions arise about the potential for relapse in Duffy-negative individuals.[36] Screening protocols may need to be adjusted to account for this risk.[37]
Our findings open up several important avenues for future research. Further studies are needed to elucidate the precise mechanisms by which P. vivax invades Duffy-negative erythrocytes. This may involve identifying alternative receptors, characterizing potential modifications to PvDBP or other parasite proteins, and investigating the role of host factors in facilitating invasion.[38] Comparative genomic and transcriptomic analyses of P. vivax isolate from Duffy-negative and Duffy-positive infections could reveal genetic adaptations that enable invasion of Duffy-negative erythrocytes.[39] This could include investigations of PvDBP gene amplifications, variations in other invasion-related genes, or changes in gene expression patterns. Studies examining the interactions between P. vivax and the host immune system in Duffy-negative individuals could provide insights into potential differences in immune responses, parasite growth, and clinical outcomes compared to infections in Duffy-positive hosts.[40] Large-scale epidemiological surveys incorporating both Duffy genotyping and sensitive P. vivax detection methods are needed to accurately assess the prevalence and distribution of P. vivax infections in Duffy-negative populations across different geographic regions.[36] Prospective clinical studies are required to evaluate the natural history, treatment responses, and potential for relapse of P. vivax infections in Duffy-negative individuals.[41] The ability of P. vivax to infect Duffy-negative erythrocytes has implications for vaccine development efforts. Vaccines targeting PvDBP may need to be reevaluated, and new vaccine candidates targeting alternative invasion pathways should be explored.[42]
Our study has a few limitations that should be acknowledged. The small sample size of Duffy-negative individuals limits the generalizability of our findings and precludes detailed statistical analyses of infection rates or risk factors. Additionally, our study design does not allow us to determine the duration or clinical course of P. vivax infections in Duffy-negative individuals, and it does not provide information on transmission dynamics or the potential for relapses.
Despite these limitations, our study has several strengths. The use of multiple complementary methods to confirm both Duffy negativity and P. vivax infection enhances the reliability of our findings. The inclusion of both molecular and microscopic detection of P. vivax provides strong evidence for true infection rather than mere exposure or PCR contamination. Furthermore, our study adds to the growing body of evidence challenging the paradigm of Duffy antigen dependency for P. vivax invasion. It highlights the need for continued research in this area.
Conclusions
In conclusion, our study provides compelling evidence for P. vivax infection in Duffy-negative individuals, challenging long-held assumptions about the relationship between P. vivax and the Duffy antigen. These findings have significant implications for our understanding of P. vivax biology, global distribution, and control strategies. They underscore the need for a re-evaluation of P. vivax epidemiology, particularly in regions with high prevalence of heterogenicity in Duffy antigens and call for further research into the mechanisms underlying this phenomenon. As we continue to strive for malaria elimination, it is crucial that we adapt our approaches to account for the evolving understanding of P. vivax infections in diverse host populations.Author Contributions
S.R. conducted the experiment. G.A. and G.K. supervised the experiments. S.R. conducted data analysis. G.A. conceptualized the project, was responsible for the overall supervision, and procured funding. S.R. wrote the manuscript. G.A. and G.K. approved the final manuscript.Acknowledgment
We are grateful to the Director General of Indian Council of Medical Research (ICMR), New Delhi, India, and the Director of ICMR-National Institute of ImmunoHematology (ICMR-NIIH), Mumbai, India, for providing funding and facilities for this study. We are thankful to Dr. Shreemati Shetty (ICMR-NIIH, Mumbai), Dr. Swati Kulkarni (ICMR-NIIH, Mumbai) and Dr. Sanmukh Joshi (Lok Samarpan Regional Blood Center, Surat) for helping in getting control and malaria positive samples.Data Availability Statement
The datasets used and/or analyzed during this study are available from the corresponding author on reasonable request.References
- Zimmerman PA. Plasmodium vivax infection in
duffy-negative people in Africa. American Journal of Tropical Medicine
and Hygiene. 2017;97:636-8. https://doi.org/10.4269/ajtmh.17-0461 PMid:28990906 PMCid:PMC5590613
- Miller
LH, Steven J. Mason, David F. Clyde, et al. The resistance factor to
Plasmodium vivax in blacks. New England Journal of Medicine.
1976;295:302-4. https://doi.org/10.1056/NEJM197608052950602 PMid:778616
- Howes
RE, Patil AP, Piel FB, et al. The global distribution of the Duffy
blood group. Nat Commun. 2011;2. doi: 10.1038/ncomms1265 https://doi.org/10.1038/ncomms1265 PMid:21468018 PMCid:PMC3074097
- Motshoge
T, Ababio GK, Aleksenko L, et al. Molecular evidence of high rates of
asymptomatic P. vivax infection and very low P. falciparum malaria in
Botswana. BMC Infect Dis. 2016;16. doi: 10.1186/s12879-016-1857-8 https://doi.org/10.1186/s12879-016-1857-8 PMid:27682611 PMCid:PMC5041318
- Mbenda
HGN, Das A. Molecular evidence of plasmodium vivax mono and mixed
malaria parasite infections in duffy-negative native cameroonians. PLoS
One. 2014;9. doi: 10.1371/journal.pone.0103262 https://doi.org/10.1371/journal.pone.0103262 PMid:25084090 PMCid:PMC4118857
- Lo
E, Yewhalaw D, Zhong D, et al. Molecular epidemiology of Plasmodium
vivax and Plasmodium falciparum malaria among duffy-positive and
Duffy-negative populations in Ethiopia. Malar J. 2015;14. doi:
10.1186/s12936-015-0596-4 https://doi.org/10.1186/s12936-015-0596-4 PMid:25884875 PMCid:PMC4340780
- Mendes
C, Dias F, Figueiredo J, et al. Duffy negative antigen is no longer a
barrier to Plasmodium vivax - molecular evidences from the African West
Coast (Angola and Equatorial Guinea). PLoS Negl Trop Dis. 2011;5. doi:
10.1371/journal.pntd.0001192 https://doi.org/10.1371/journal.pntd.0001192 PMid:21713024 PMCid:PMC3119644
- Niangaly
A, Gunalan K, Ouattara A, et al. Plasmodium vivax Infections over 3
Years in Duffy Blood Group Negative Malians in Bandiagara, Mali.
American Journal of Tropical Medicine and Hygiene. 2017;97:744-52. doi:
10.4269/ajtmh.17-0254 https://doi.org/10.4269/ajtmh.17-0254 PMid:28749772 PMCid:PMC5590610
- Wurtz N, Lekweiry KM, Bogreau H, et al. Vivax malaria in Mauritania includes infection of a Duffy-negative individual. 2011. https://doi.org/10.1186/1475-2875-10-336 PMid:22050867 PMCid:PMC3228859
- Niang
M, Thiam LG, Sow A, et al. A molecular survey of acute febrile
illnesses reveals Plasmodium vivax infections in Kedougou, southeastern
Senegal. Malar J. 2015;14. doi: 10.1186/s12936-015-0808-y https://doi.org/10.1186/s12936-015-0808-y PMid:26186936 PMCid:PMC4506577
- Abdelraheem
MH, Albsheer MMA, Mohamed HS, et al. Transmission of Plasmodium vivax
in Duffy-negative individuals in central Sudan. Trans R Soc Trop Med
Hyg. 2016;110:258-60. doi: 10.1093/trstmh/trw014 https://doi.org/10.1093/trstmh/trw014 PMid:27076512
- Asua
V, Tukwasibwe S, Conrad M, et al. Plasmodium species infecting children
presenting with malaria in Uganda. American Journal of Tropical
Medicine and Hygiene. 2017;97:753-7. doi: 10.4269/ajtmh.17-0345 https://doi.org/10.4269/ajtmh.17-0345 PMid:28990911 PMCid:PMC5590612
- Ménard
D, Barnadas C, Bouchier C, et al. Plasmodium vivax clinical malaria is
commonly observed in Duffy-negative Malagasy people. Proc Natl Acad Sci
U S A. 2010;107:5967-71. doi: 10.1073/pnas.0912496107 https://doi.org/10.1073/pnas.0912496107 PMid:20231434 PMCid:PMC2851935
- Cavasini
CE, De Mattos LC, Couto ÁARDA, et al. Duffy blood group gene
polymorphisms among malaria vivax patients in four areas of the
Brazilian Amazon region. Malar J. 2007;6. doi: 10.1186/1475-2875-6-167 https://doi.org/10.1186/1475-2875-6-167 PMid:18093292 PMCid:PMC2244634
- Barbosa
S, Gozze AB, Lima NF, et al. Epidemiology of Disappearing Plasmodium
vivax Malaria: A Case Study in Rural Amazonia. PLoS Negl Trop Dis.
2014;8. doi: 10.1371/journal.pntd.0003109 https://doi.org/10.1371/journal.pntd.0003109 PMid:25166263 PMCid:PMC4148206
- Hostetler
JB, Lo E, Kanjee U, et al. Independent Origin and Global Distribution
of Distinct Plasmodium vivax Duffy Binding Protein Gene Duplications.
PLoS Negl Trop Dis. 2016;10:e0005091. doi: 10.1371/journal.pntd.0005091
https://doi.org/10.1371/journal.pntd.0005091 PMid:27798646 PMCid:PMC5087946
- Russo
G, Faggioni G, Paganotti GM, et al. Molecular evidence of Plasmodium
vivax infection in Duffy negative symptomatic individuals from Dschang,
West Cameroon. Malar J. 2017;16:1-9. doi: 10.1186/s12936-017-1722-2 https://doi.org/10.1186/s12936-017-1722-2 PMid:28196496 PMCid:PMC5309988
- Kahar
MA, Patel RD. Phenotype frequencies of blood group systems (Rh, Kell,
Kidd, Duffy, MNS, P, Lewis, and Lutheran) in blood donors of south
Gujarat, India. Asian J Transfus Sci. 2014;8:51-5. doi:
10.4103/0973-6247.126693 https://doi.org/10.4103/0973-6247.126693 PMid:24678176 PMCid:PMC3943149
- Das
MK, Singh SS, Adak T, et al. The Duffy blood groups of Jarawas - The
primitive and vanishing tribe of Andaman and Nicobar Islands of India.
Transfusion Medicine. 2005;15:237-40. doi:
10.1111/j.1365-3148.2005.00583.x https://doi.org/10.1111/j.1365-3148.2005.00583.x PMid:15943709
- WHO. Basic malaria microscopy. WHO 2010.
- WHO. Malaria parasite counting, malaria microscopy. In: WHO, ed. Standard operating procedure-MM-SOP-09. 2016.
- Shaikh
R, Kanjaksha G, Kashivishwanath V, et al. ACKR1 gene polymorphisms in
Bombay blood group (Oh) individuals of Indian origin. Transfusion and
Apheresis Science. 2024;63. doi: 10.1016/j.transci.2024.103975 https://doi.org/10.1016/j.transci.2024.103975 PMid:39126827
- Zimmerman
PA, Ferreira MU, Howes RE, et al. Red Blood Cell Polymorphism and
Susceptibility to Plasmodium vivax. Advances in Parasitology. Academic
Press 2013:27-76. https://doi.org/10.1016/B978-0-12-407826-0.00002-3 PMid:23384621 PMCid:PMC3728992
- Tournamille
C, Colin Y, Cartron JP, et al. Disruption of a GATA motif in the Duffy
gene promoter abolishes erythroid gene expression in Duffy-negative
individuals. Nature Genetics 1995 10:2. 1995;10:224-8. doi:
10.1038/ng0695-224 https://doi.org/10.1038/ng0695-224 PMid:7663520
- Iwamoto
S, Li J, Omi T, et al. Identification of a Novel Exon and Spliced Form
of Duffy mRNA That Is the Predominant Transcript in Both Erythroid and
Postcapillary Venule Endothelium Blood. 1996 Jan 1;87(1):378-85. https://doi.org/10.1182/blood.V87.1.378.bloodjournal871378 PMid:8547665
- Langhi DM, Bordin JO. Duffy blood group and malaria. Hematology. 2006;11:389-98. doi: 10.1080/10245330500469841 https://doi.org/10.1080/10245330500469841 PMid:17607593
- Adams
JH, Hudson DE, Torii M, et al. The Duffy Receptor Family of Plasmodium
knowlesi Is Located within the Micronemes of Invasive Malaria
Merozoites. 1990. https://doi.org/10.1016/0092-8674(90)90295-P PMid:2170017
- Gruszczyk
J, Kanjee U, Chan LJ, et al. Transferrin receptor 1 is a
reticulocyte-specific receptor for Plasmodium vivax. Science (1979).
2018;359:48-55. doi: 10.1126/science.aan1078 https://doi.org/10.1126/science.aan1078 PMid:29302006 PMCid:PMC5788258
- Gruszczyk
J, Huang RK, Chan LJ, et al. Cryo-EM structure of an essential
Plasmodium vivax invasion complex. Nature. 2018;559:135-9. doi:
10.1038/s41586-018-0249-1 https://doi.org/10.1038/s41586-018-0249-1 PMid:29950717
- Gunalan
K, Lo E, Hostetler JB, et al. Role of Plasmodium vivax Duffy-binding
protein 1 in invasion of Duffy-null Africans. Proc Natl Acad Sci U S A.
2016;113:6271-6. doi: 10.1073/pnas.1606113113 https://doi.org/10.1073/pnas.1606113113 PMid:27190089 PMCid:PMC4896682
- Abagero
BR, Rama R, Obeid A, et al. Detection of Duffy blood group genotypes
and submicroscopic Plasmodium infections using molecular diagnostic
assays in febrile malaria patients. Malar J. 2024;23. doi:
10.1186/s12936-024-04875-5 https://doi.org/10.1186/s12936-024-04875-5 PMid:38902674 PMCid:PMC11191254
- Malleret
B, Li A, Zhang R, et al. Plasmodium vivax: restricted tropism and rapid
remodeling of CD71-positive reticulocytes. Blood. 2015;125:1314-24.
doi: 10.1182/blood-2014 https://doi.org/10.1182/blood-2014-08-596015 PMid:25414440 PMCid:PMC4401350
- Culleton R, Carter R. African Plasmodium vivax: Distribution and origins. Int J Parasitol. 2012;42:1091-7. https://doi.org/10.1016/j.ijpara.2012.08.005 PMid:23017235
- Howes
RE, Reiner RC, Battle KE, et al. Plasmodium vivax Transmission in
Africa. PLoS Negl Trop Dis. 2015;9. doi: 10.1371/journal.pntd.0004222 https://doi.org/10.1371/journal.pntd.0004222 PMid:26587988 PMCid:PMC4654493
- Gething
PW, Elyazar IRF, Moyes CL, et al. A Long Neglected World Malaria Map:
Plasmodium vivax Endemicity in 2010. PLoS Negl Trop Dis. 2012;6. doi:
10.1371/journal.pntd.0001814 https://doi.org/10.1371/journal.pntd.0001814 PMid:22970336 PMCid:PMC3435256
- Twohig
KA, Pfeffer DA, Baird JK, et al. Growing evidence of Plasmodium vivax
across malaria-endemic Africa. PLoS Negl Trop Dis. 2019;13. doi:
10.1371/journal.pntd.0007140 https://doi.org/10.1371/journal.pntd.0007140 PMid:30703083 PMCid:PMC6372205
- Battle
KE, Lucas TCD, Nguyen M, et al. Mapping the global endemicity and
clinical burden of Plasmodium vivax, 2000-17: a spatial and temporal
modelling study. The Lancet. 2019;394:332-43. doi:
10.1016/S0140-6736(19)31096-7 https://doi.org/10.1016/S0140-6736(19)31096-7 PMid:31229233
- Ntumngia
FB, Thomson-Luque R, de Menezes Torres L, et al. A novel erythrocyte
binding protein of Plasmodium vivax suggests an alternate invasion
pathway into duffy-positive reticulocytes. mBio. 2016;7. doi:
10.1128/mBio.01261-16 https://doi.org/10.1128/mBio.01261-16 PMid:27555313 PMCid:PMC4999553
- Hupalo
DN, Luo Z, Melnikov A, et al. Population genomics studies identify
signatures of global dispersal and drug resistance in Plasmodium vivax.
Nat Genet. 2016;48:953-8. doi: 10.1038/ng.3588 https://doi.org/10.1038/ng.3588 PMid:27348298 PMCid:PMC5347536
- Kano
FS, Sanchez BAM, Sousa TN, et al. Plasmodium vivax Duffy binding
protein: Baseline antibody responses and parasite polymorphisms in a
well-consolidated settlement of the Amazon Region. Tropical Medicine
and International Health. 2012;17:989-1000. doi:
10.1111/j.1365-3156.2012.03016.x https://doi.org/10.1111/j.1365-3156.2012.03016.x PMid:22643072
- Bassat
Q, Velarde M, Mueller I, et al. Key knowledge gaps for Plasmodium vivax
control and elimination. American Journal of Tropical Medicine and
Hygiene. 2016;95:62-71. https://doi.org/10.4269/ajtmh.16-0180 PMid:27430544 PMCid:PMC5201224
- Beeson
JG, Kurtovic L, Dobaño C, et al. Challenges and strategies for
developing efficacious and long-lasting malaria vaccines. 2019. https://doi.org/10.1126/scitranslmed.aau1458 PMid:30626712