The prevalence of EBV ranges from 50% in children to 90% in adults worldwide.[4] In developing countries, exposure typically occurs early in life, while the onset of infection is delayed in areas with greater socioeconomic development.[3] EBV is primarily transmitted by infected saliva, which promotes access and entry into the upper respiratory tree's epithelial cells and B lymphocytes as the main target cells; EBV also spreads through breast milk, body fluid, blood transfusion, and transplantation.[5,6] Similar to other herpes viruses, its life cycle is characterized by a lytic phase where it can infect other cells or spread the infection to other individuals, and the more quiescent latent phase where it persists lifelong in memory B cells.[7]
EBV genome is 172 kbp in size and encodes more than 85 genes, which express proteins depending on the viral life cycle phase.[8] Two EBV subtypes can infect humans: EBV1 and EBV2. The biological characteristics of the two genotypes differ; EBV genotype 1 is more effective in immortalizing B cells, while EBV genotype 2 has a higher lytic ability, these types differ in their EBV nuclear antigens: (EBNA-2) and (EBNA-3) sequences (EBNA-3a, EBNA-3b, and EIBNA-3c).[9] The most studied and approved method used to differentiate the two types is EBNA3C.[10]
Infected with EBV in healthy individuals is well controlled by the immune system cells, including cytotoxic T-cells and natural killer cells, making the lytic phase of infection recede and latent infection of the memory B lymphocytes predominant.[11] Primary infection in most immune-competent individuals is usually asymptomatic, mainly in infants and childhood, or causes infectious mononucleosis disease (IM) in adolescents or young adults with unspecific symptoms like fever, malaise, pharyngitis, and lymphadenopathy.[12]
However, under immunocompromised conditions, as in hematopoietic stem cell transplant recipients (HSCT), who undergo these transplants as crucial treatments for various malignancies and immune deficiency diseases, EBV primary infection and reactivation are frequent complications. The risk of infection increases when using immunosuppressive regimens that prevent graft rejection.[13] These agents inhibit cytotoxic T-cells, which are crucial for killing infected B-cells. In some cases, the continuous use of these conditioning regimens increases the opportunity to proliferate infected B-cells, resulting in EBV-related post-transplant lymphoproliferative disorders (PTLDs), which are life-threatening complications that facilitate tumorigenesis.[14] Therefore, the evolution of immunosuppression for transplantation has reduced the incidence of rejection but has increased the risk of infection and virally mediated malignancies.[15]
Previous studies indicated a predominance of EBV 1 strains.[16] More data suggest that a notable proportion of individuals are infected with EBV type 2 and that co-infection of both types, EBV1 and EBV2, is more frequently found in immunocompromised individuals.[17,18]
To the best of our knowledge, this is the first study that examined the genotypes of EBV in transplant patients. Therefore, this study aimed to define EBV genotyping among HSCT recipients in Jordan and to determine the association between EBV positivity and demographic factors, including gender and age.
Materials and Methods
The Study Design and Population. A retrospective observational study was conducted from January to October 2024 to characterize EBV in patients who underwent HSCT at the Jordanian Royal Medical Services Hospital (JRMS) post-transplantation. The study included all patients aged one year and older who had received HSCT, regardless of gender. However, patients with significant medical conditions that could impact the study outcomes, such as organ failure at the time of transplantation, were excluded from the analysis.Sample Collection. Blood samples were collected in EDTA tubes, centrifuged for 10 minutes at 3000 rpm, and stored as plasma at -20°C for further processing.
DNA Extraction. Following the manufacturer’s protocol, DNA was isolated from 200 μL of plasma using QIAmap DNA Mini Kit (Qiagen, Hilden, Germany). Extracted DNA samples were measured for purity and concentration using the NanoDrop-2000 (Thermo Fisher Scientific, Waltham, Massachusetts) spectrophotometry. Purity was evaluated by the absorbance ratio at 260/280 nm, with values between (1.8 and 2.0) considered indicative of high purity. DNA concentrations were quantified at 260 nm and recorded in ng/μL. Isolated DNA samples were subsequently stored at -20°C until further use.[19]
EBV DNA detection by Real-time PCR. The EBV DNA detection and viral load quantitation were detected using the Artus® EBV RG PCR Kit (Qiagen, Hilden, Germany) on the Rotor-Gene Q instrument using 20 μL of DNA following the manufacturer’s instructions. This detection method is based on real-time PCR amplification using a 30μL EBV RG master mix that contains primers, enzymes, and probes designed to target regions within the EBNA-1 gene.[10] The fluorescence reporter dyes were used for direct detection of the amplified product. Thermal cycling conditions were as follows: initial denaturation at 95°C for 10 minutes, followed by 45 cycles of amplification with denaturation at 95°C for 15 seconds, lowered to 65°C annealing temperature for 30 seconds, extended at 72°C for 20 seconds.[20]
EBV Genotyping by Conventional PCR of the EBNA3C gene. Epstein-Barr virus genotyping was performed using Conventional PCR techniques, specifically targeting the EBNA3C gene, which serves as a crucial marker for differentiating between the various EBV types based on the size of the PCR products generated: a product size of 153 base pairs (bp) indicates EBV type 1, whereas a product size of 246 (bp) correspond to EBV type 2. The specific primer sequences employed in this amplification process were as follows: the forward primer EBNA3C1 was 5′-GCCAGAGGTAAGTGGACTTT-3′, and the reverse primer EBNA3C2 was 5′-TGGAGAGGTCAGGTTACTTA-3′.[21]
PCR amplification was performed using the Quant Gene 9600 instrument (Bioer, China). The PCR reaction mixture was performed in 20 µL total volume consisting of 10 µL of 2X PCR Master Mix Solution (i-pfu, iNtRON Biotechnology, South Korea), 2 µL of extracted DNA, 1 µL of forward primer (10 µM), 1 µL of reverse primer (10 µM), and 6 µL of nuclease-free water to achieve the desired volume. The PCR thermal cycling conditions were as follows: initial denaturation at 94° for 2 minutes, 40 cycles of: Denaturation at 94°C for 30 seconds, Annealing at 56°C for 30 seconds, and extension at 72°C for 1 minute, the final extension at 72°C for 5 minutes. Nuclease-free water was used as a negative control to ensure the validity of the result.[21]
Following PCR, the amplified DNA fragments underwent separation through electrophoresis using 2% UltraPhore agarose gel (Condalab, Spain). The gel matrix was prepared, dissolving the agarose in Tris-acetate-EDTA (TAE) buffer; then 5 µL of RedSafe dye was incorporated to enhance visualization by binding to the DNA bands and making fluoresce under the UV light. The PCR products were loaded into the gel wells alongside the SiZer™-100 DNA Marker Solution (iNtRON Biotechnology) and the ZR 50 bp DNA Marker (Zymo Research), reference standards to estimate the PCR product size. Electrophoresis was conducted at 90V for approximately 60 minutes. After completion of the electrophoresis run, the gel was subjected to UV light exposure using a transilluminator and a Quantum gel documentation system (Vilber, France) for DNA band visualizing. Band sizes of 153 bp and 246 bp were used to confirm the presence of EBV1 and EBV2, respectively.[21]
Statistical Analysis. Statistical analysis was performed using IBM SPSS statistical software version 25. Qualitative data were presented as frequencies and percentages. Quantitative data included were expressed as mean ± standard deviation (SD). The association between categorical variables was assessed using The Chi-square. The P-values < 0.05 were considered statistically significant.
Ethical Consideration. This retrospective observational study used ethical guidelines and principles from the Declaration of Helsinki. It was approved by the Institutional Review Board at Zarqa University (IRB/ZU/2024/29). This study was designed to minimize any potential risks to participants. All procedures involving human participants followed the institution’s standards for ethical research. Additionally, all collected data were anonymized prior to analysis to protect participant privacy.
Results
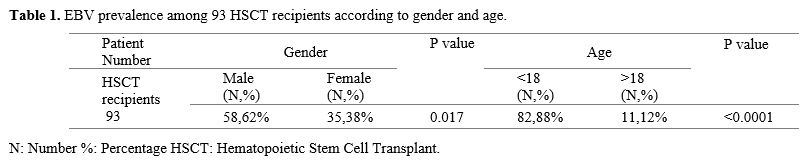
Distribution of EBV among HSCT recipients according to Gender and Age. Among 93 EBV-positive HSCT recipients, 58 (62%) were males, and 35 (38%) were females, showing a higher significance of EBV prevalence in males (P<0.017). Regarding age, 82 (88%) of the recipients were under 18 years, while only 11 (12%) were above 18 years, having a significantly higher EBV prevalence in ages under 18 years (P<0.0001), as shown in Table 1.EBV-Genotypes prevalence among HSCT according to gender and age. Out of the 93 EBV-positive samples, 34 were analyzed for genotyping, as the remaining samples were insufficient for analysis. Three of the 34 positive samples (1, 13, and 25) showed no bands on the gel electrophoresis.
The distribution of EBV genotypes among the 31 recipients of HSCT was analyzed, with results categorized by gender and age. Among the 31 EBV-positive samples, 26 patients (84%) were found to have genotype 2, whereas five patients (16%) had mixed infections involving both EBV genotype 1 and genotype 2. These results demonstrate that EBV genotype 2 is the most prevalent strain within the HSCT sample population studied (P<0.0001).
According to gender, from 26 positive EBV2 samples, 15 were males (58%), and 11 (42%) were females. No significant difference in EBV2 prevalence between genders (p=0.43). Of the co-infection samples, all were males. Among the 26 positive EBV2 samples, 24 patients were under 18 years (92%) while only 2 patients were above years 18 (8%) and this showed a significant difference (P <0.0001), Among the age groups. In contrast, 3 patients of mixed EBV type were under 18 years, and 2 patients were above 18 years, as shown in Table 2.
 |
|
Patient characteristics. The distribution of age among the EBV-positive samples comprised a total of 34 patients, categorized into seven age groups, with the majority falling within the (1-5) year age group, accounting for 21 patients (61.76%). The next largest group was the 6 to 10-year age range, comprising 7 patients (20.58%), followed by the 11 to 15-year group with 4 patients (11.76%). Notably, there were no patients in the age groups 16-20 and 21-25 years. The age group of 26-30 years included 1 patient (2.94%), as did the group over 30 years. The mean age of the patients was 6.88±7.09 SD.
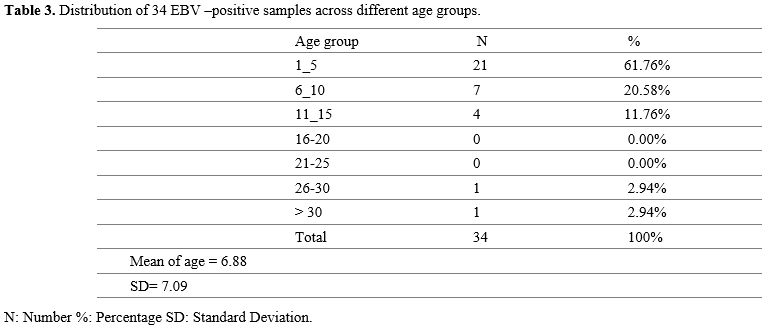
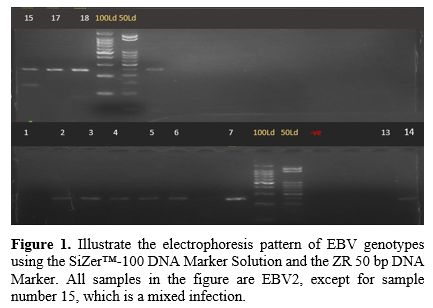
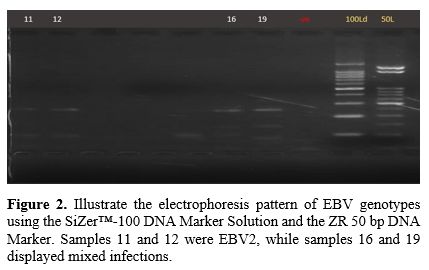
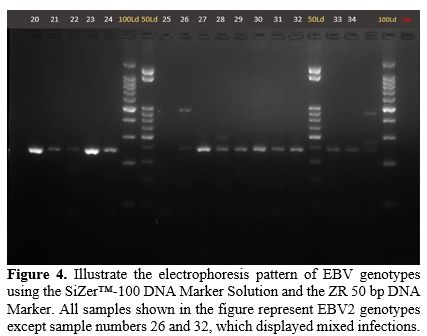
Gel Electrophoresis Results. Figures (Figure 1-4) illustrate the electrophoresis pattern of EBV genotypes using the SiZer™-100 DNA Marker Solution and the ZR 50 bp DNA Marker.
 |
Figure 3.
Illustrate the electrophoresis pattern of EBV genotypes using the
SiZer™-100 DNA Marker Solution and the ZR 50 bp DNA Marker. All samples
shown in the figure represent EBV2 genotypes. |
Discussion
Hematopoietic stem cell transplantation is the pivotal treatment for patients with hematological malignancies. However, the extensive immunosuppressive regimens required to prevent graft rejection and support the engraftment process increase the risk of infection.[22] The EBV virus poses a substantial risk to immunocompromised recipients because of its potential to reactivate, leading to life-threatening malignancies if not properly managed with appropriate therapy.[23]Differences in the EBNA3C gene have been documented as a basis for distinguishing EBV types in the current study. The finding in the 31 HSCT recipients samples showed that genotype 2 was identified in 26 samples (84%), and five recipients (16%) demonstrated a mixed infection of EBV1+ EBV2. This finding suggests that EBV2 is the predominant genotype within the studied population. Compared to other studies conducted for different immunocompromised statuses, our findings indicate a higher prevalence of EBV2. Specifically, Ayee et al.,[19] reported that (52%) of EBV2 was predominant among NCP patients in Ghana and Palma et al.,[24] observed a prevalence of EBV2 with (47.6%) in pediatric HL patients and (69.2%) in adult HL patients in Mexico. In comparison to studies conducted in the Middle East, EBV genotype 1 is revealed across various populations, (72.5%) among healthy blood donors in Qatar,[25] (100%) in lymphoma patients in Yemen,[26] and (91.2%) in patients with hematologic malignancies in Iran.[27] These disparities in EBV-genotype prevalence could be mainly due to the geographic and demographic variations, the immune status of the populations within each country, and the sample size for each study.
EBV mixed infection was identified in (16%) of cases. Many genotype studies conducted for patients with Human Immunodeficiency virus (HIV) who are also immunocompromised have a comparable with our finding with lower co-infection rate in Ethiopia (4.7%)[10] and China (12.96%);[28] on the contrary, Brazil demonstrates a higher prevalence rate (26.32%).[29] We did not observe any significant differences based on gender, whereas age was significantly associated with EBV2 in individuals under 18 years old. As seen in a study conducted in Brazil, IM patients recorded a much higher rate of EBV2 for age within 10-15 years (62.5%) compared to EBV1 (25%).[21]
A significant difference in EBV infection was observed in males (62%) more than in females (38%), and a higher significance was observed in recipients under 18 years (88%) more than in recipients over 18 years (12%). Most studies did not find a significant age difference[30,31] or gender difference[32,33] as a risk for infection. However, some studies revealed that EBV DNAemia is detected more frequently in immunocompromised children (12.6%) compared to adults (6.2%), emphasizing their vulnerability to primary or reactivated infections post-transplant.[34]
Like other retrospective studies, the current study faced limitations such as gaps in record keeping and limited access to data. Additionally, the small sample size was drawn from only one hospital. We recommend conducting further research, including a broader range of data and a larger sample size, and performing whole genome sequencing to provide a clearer understanding of the diversity of EBV strains in HSCT patients.
Conclusions
This study represents the first investigation into the prevalence of EBV genotypes among HSCT recipients in Jordan. The findings indicate that the predominant genotype identified in this patient population is EBV 2, followed by mixed infections involving both EBV 1 and EBV 2. Moreover, a significant association was observed between EBV positivity and male recipients. Additionally, the prevalence of EBV was notably higher among recipients aged under 18 years. Identifying the most common genotypes in transplant patients enhances the treatment management of complications associated with EBV.Acknowledgments
We sincerely appreciate the financial support provided by Zarqa University, which made this research possible.References
- Young, L.S., L.F. Yap, and P.G. Murray,
Epstein-Barr virus: more than 50 years old and still providing
surprises. Nature reviews cancer, 2016. 16(12): p. 789-802. https://doi.org/10.1038/nrc.2016.92 PMid:27687982
- Farrell, P.J., Epstein-Barr virus and cancer. Annual Review of Pathology: Mechanisms of Disease, 2019. 14(1): p. 29-53. https://doi.org/10.1146/annurev-pathmechdis-012418-013023 PMid:30125149
- Gewurz, B., R. Longnecker, and J. Cohen, Epstein-barr virus. Fields Virology, 2021. 2: p. 7.
- Styczynski,
J., et al., Management of Epstein-Barr Virus infections and
post-transplant lymphoproliferative disorders in patients after
allogeneic hematopoietic stem cell transplantation: Sixth European
Conference on Infections in Leukemia (ECIL-6) guidelines.
Haematologica, 2016. 101(7): p. 803. https://doi.org/10.3324/haematol.2016.144428 PMid:27365460 PMCid:PMC5004459
- Tzellos,
S., et al., A single amino acid in EBNA-2 determines superior B
lymphoblastoid cell line growth maintenance by Epstein-Barr virus type
1 EBNA-2. Journal of virology, 2014. 88(16): p. 8743-8753. https://doi.org/10.1128/JVI.01000-14 PMid:24850736 PMCid:PMC4136291
- Damania,
B., S.C. Kenney, and N. Raab-Traub, Epstein-Barr virus: Biology and
clinical disease. Cell, 2022. 185(20): p. 3652-3670. https://doi.org/10.1016/j.cell.2022.08.026 PMid:36113467 PMCid:PMC9529843
- Hislop,
A.D., et al., Cellular responses to viral infection in humans: lessons
from Epstein-Barr virus. Annu. Rev. Immunol., 2007. 25: p. 587-617. https://doi.org/10.1146/annurev.immunol.25.022106.141553 PMid:17378764
- Smatti,
M.K., et al., Epstein-Barr virus epidemiology, serology, and genetic
variability of LMP-1 oncogene among healthy population: an update.
Frontiers in oncology, 2018. 8: p. 211. https://doi.org/10.3389/fonc.2018.00211 PMid:29951372 PMCid:PMC6008310
- Tzellos, S. and P.J. Farrell, Epstein-Barr virus sequence variation-biology and disease. Pathogens, 2012. 1(2): p. 156-175. https://doi.org/10.3390/pathogens1020156 PMid:25436768 PMCid:PMC4235690
- Zealiyas,
K., et al., Genotype characterization of Epstein-Barr virus among
adults living with human immunodeficiency virus in Ethiopia. Frontiers
in Microbiology, 2023. 14: p. 1270824. https://doi.org/10.3389/fmicb.2023.1270824 PMid:38029140 PMCid:PMC10644458
- Martinez,
O.M., The Biology of Epstein-Barr Virus and Posttransplant
Lymphoproliferative Disease. Post-Transplant Lymphoproliferative
Disorders, 2010: p. 29-43. https://doi.org/10.1007/978-3-642-01653-0_4 PMCid:PMC2848143
- Crawford,
D.H., et al., A cohort study among university students: identification
of risk factors for Epstein-Barr virus seroconversion and infectious
mononucleosis. Clinical infectious diseases, 2006. 43(3): p. 276-282. https://doi.org/10.1086/505400 PMid:16804839
- Green,
M. and M. Michaels, Epstein-Barr virus infection and posttransplant
lymphoproliferative disorder. American Journal of Transplantation,
2013. 13: p. 41-54. https://doi.org/10.1111/ajt.12004 PMid:23347213
- Landgren,
O., et al., Risk factors for lymphoproliferative disorders after
allogeneic hematopoietic cell transplantation. Blood, The Journal of
the American Society of Hematology, 2009. 113(20): p. 4992-5001. https://doi.org/10.1182/blood-2008-09-178046 PMid:19264919 PMCid:PMC2686146
- Ciancio,
G., G.W. Burke, and J. Miller, Induction therapy in renal
transplantation: an overview of current developments. Drugs, 2007. 67:
p. 2667-2680. https://doi.org/10.2165/00003495-200767180-00003 PMid:18062717
- Zanella,
L., et al., A reliable Epstein-Barr Virus classification based on
phylogenomic and population analyses. Scientific reports, 2019. 9(1):
p. 9829. https://doi.org/10.1038/s41598-019-45986-3 PMid:31285478 PMCid:PMC6614506
- Chang,
C.M., et al., The extent of genetic diversity of Epstein-Barr virus and
its geographic and disease patterns: a need for reappraisal. Virus
research, 2009. 143(2): p. 209-221. https://doi.org/10.1016/j.virusres.2009.07.005 PMid:19596032 PMCid:PMC2731007
- Neves,
M., et al., Epstein-Barr virus strains and variations: Geographic or
disease‐specific variants? Journal of medical virology, 2017. 89(3): p.
373-387. https://doi.org/10.1002/jmv.24633 PMid:27430663
- Ayee,
R., et al., Genotypic characterization of Epstein Barr virus in blood
of patients with suspected nasopharyngeal carcinoma in Ghana. Viruses,
2020. 12(7): p. 766. https://doi.org/10.3390/v12070766 PMid:32708700 PMCid:PMC7412455
- Miller,
J.A., et al., Comparison of Real-Time PCR and Digital PCR for Detection
of Plasma Epstein-Barr Virus DNA in Nasopharyngeal Carcinoma. The
Journal of Molecular Diagnostics, 2023. 25(7): p. 490-501. https://doi.org/10.1016/j.jmoldx.2023.03.007 PMid:37068736
- Monteiro,
T.A.F., et al., Genotypes of Epstein-Barr virus (EBV1/EBV2) in
individuals with infectious mononucleosis in the metropolitan area of
Belém, Brazil, between 2005 and 2016. Brazilian Journal of Infectious
Diseases, 2020. 24(4): p. 322-329. https://doi.org/10.1016/j.bjid.2020.06.004 PMid:32619403 PMCid:PMC9392146
- Henig,
I. and T. Zuckerman, Hematopoietic stem cell transplantation-50 years
of evolution and future perspectives. Rambam Maimonides medical
journal, 2014. 5(4). https://doi.org/10.5041/RMMJ.10162 PMid:25386344 PMCid:PMC4222417
- Tsushima,
T., et al., Clinical characteristics and outcomes of Epstein-Barr virus
viral load after allogeneic hematopoietic stem cell transplantation.
Annals of Hematology, 2024. 103(3): p. 935-946. https://doi.org/10.1007/s00277-023-05596-6 PMid:38157001 PMCid:PMC10867052
- Palma,
I., et al., Detection of Epstein-Barr virus and genotyping based on
EBNA2 protein in Mexican patients with Hodgkin lymphoma: a comparative
study in children and adults. Clinical Lymphoma Myeloma and Leukemia,
2013. 13(3): p. 266-272. https://doi.org/10.1016/j.clml.2012.11.010 PMid:23276887
- Smatti,
M.K., et al., Prevalence and molecular profiling of Epstein Barr virus
(EBV) among healthy blood donors from different nationalities in Qatar.
PLoS One, 2017. 12(12): p. e0189033. https://doi.org/10.1371/journal.pone.0189033 PMid:29228016 PMCid:PMC5724864
- Al-Mahbashi,
A.A., et al., Epstein-Barr Virus Genotypes and Phylogeny among Cancer
Patients in Sana'a City, Yemen. Jordan Journal of Biological Sciences,
2023. 16(1). https://doi.org/10.54319/jjbs/160118
- Tabibzadeh,
A., et al., Molecular epidemiology of epstein-barr virus (ebv) in
patients with hematologic malignancies. Asian Pacific journal of cancer
prevention, 2020. 21(3): p. 693-698. https://doi.org/10.31557/APJCP.2020.21.3.693 PMid:32212795 PMCid:PMC7437315
- Wan,
Z., et al., Epstein-Barr virus variation in people living with human
immunodeficiency virus in southeastern China. Virology Journal, 2023.
20(1): p. 107. https://doi.org/10.1186/s12985-023-02078-z PMid:37259131 PMCid:PMC10230783
- Pereira,
L.M.S., et al., Epstein-Barr Virus (EBV) Genotypes Associated with the
Immunopathological Profile of People Living with HIV-1: Immunological
Aspects of Primary EBV Infection. Viruses, 2022. 14(2): p. 168. https://doi.org/10.3390/v14020168 PMid:35215762 PMCid:PMC8880155
- Patriarca,
F., et al., Prognostic factors and outcome of E pstein-B arr virus DNA
emia in high‐risk recipients of allogeneic stem cell transplantation
treated with preemptive rituximab. Transplant Infectious Disease, 2013.
15(3): p. 259-267. https://doi.org/10.1111/tid.12061 PMid:23405972
- Marinho‑Dias,
J., et al., Association of Epstein‑Barr virus infection with allogeneic
hematopoietic stem cell transplantation in patients in Portugal.
Molecular Medicine Reports, 2019. 19(3): p. 1435-1442. https://doi.org/10.3892/mmr.2018.9794 PMCid:PMC6390016
- Burns,
D.M., et al., Greatly reduced risk of EBV reactivation in
rituximab-experienced recipients of alemtuzumab-conditioned allogeneic
HSCT. Bone marrow transplantation, 2016. 51(6): p. 825-832. https://doi.org/10.1038/bmt.2016.19 PMid:26901708 PMCid:PMC4880046
- Sanz,
J., et al., EBV-associated post-transplant lymphoproliferative disorder
after umbilical cord blood transplantation in adults with hematological
diseases. Bone marrow transplantation, 2014. 49(3): p. 397-402. https://doi.org/10.1038/bmt.2013.190 PMid:24292521
- Gupta,
S., et al., Clinical utility of EBV DNA testing of blood in
immunocompetent and immunocompromised patients: A single-center
experience. American Journal of Clinical Pathology, 2024: p. aqae143. https://doi.org/10.1093/ajcp/aqae143 PMid:39450750