Tumor necrosis factor receptor-associated factor.[6] (TRAF6) is an E3 ubiquitin ligase and an adaptor molecule in tumor necrosis factor receptor-mediated signaling pathways.[8,9] Of note, TRAF6 is found to be ectopically expressed in 59.6% of all tumors, including uterine fibroids, colon, gastric, breast, and glioma tumors, and affects cancer progression by regulating cancer cell proliferation, apoptosis, invasion, and migration.[10,11] Increasing evidence has demonstrated that TRAF6 activates the inflammatory and pyroptotic responses of NK/T cells, thus disturbing the homeostasis of NK/T cells.[12,13] Interestingly, TRAF6 can potentiate the malignant behaviors of NKTL cells by acting as an activator of nuclear factor κB (NF-κB).[14] Although previous studies have mainly focused on the oncogenic role of TRAF6 in B cell lymphoma,[15-17] its role in NKTL remains poorly understood.
Ubiquitination is a ubiquitin ligase-mediated posttranslational modification of proteins to target them for proteasomal degradation or non-degradative signaling, leading to alterations in tumor-suppressing and tumor-promoting proteins in cancer.[18] TRAF6 catalyzes ubiquitination to degrade cancer-related proteins to regulate cancer progression.[19-21] As known before, macrophage stimulating 1 (MST1) is a core component of the Hippo pathway that plays a role in the anti-cancer immune response.[22] TRAF6 can bind to MST1 and degrade it via ubiquitination.[23] More importantly, MST1 is noted to be downregulated in NKTL tissues and acts as a suppressor of NKTL cell growth,[24] which provides the feasibility of exploring the downstream mechanism of TRAF6 in NKTL cells.
Taking the above evidence into consideration, we speculated that TRAF6 plays a role in NKTL cell proliferation via ubiquitination-mediated degradation of MST1. Consequently, our study is conducted to analyze the molecular mechanism of TRAF6 in the malignant proliferation of NKTL cells and offer a novel theoretical basis for the treatment of NKTL.
Methods and materials
Cell culture and treatment. ANK1 cells were provided by Shanghai Yaji Biological Co., Ltd (Shanghai, China), KHYG-1 cells were obtained from the JCRB cell bank (Tokyo, Japan), and SNK-6 and normal NK cells were provided by American Type Culture Collection (ATCC, Manassas, VA, USA). HANK1 cells were inoculated in Iscove's modified Dulbecco medium (Gibco, Grand Island, NY, USA) containing 20% fetal bovine serum (FBS) and 100 U/mL interleukin-2 (IL-2). SNK-6 and normal NK cells were inoculated in Roswell Park Memorial Institute medium-1640 (RPMI-1640, Gibco) containing 10% FBS and 700 U/mL IL-2. KHYG-1 cells were inoculated in RPMI-1640 medium supplemented with 10% FBS and 100 U/mL IL-2. All cells were cultured under an environment of 37℃ and 5% CO2.Cell transfection. Three strands of small interfering RNAs (siRNAs) targeting TRAF6 (si-TRAF6#1, si-TRAF6#2, and si-TRAF6#3), three strands of siRNAs targeting MST1 (si-MST1#1, si-MST1#2, and si-MST1#3), and their negative control (si-NC) were procured from Gene Pharma (Shanghai, China) and transfected into HANK1 cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) for 48 h before the subsequent experiments.
Cell counting kit-8 (CCK-8) method. HANK1 cell proliferation was evaluated using the CCK-8 method. Transfected HANK1 cells were seeded into the 96-well plate (1 × 104 cells/well) and cultured at 37 °C with 5% CO2. On days 1, 2, and 3 of incubation, cells in each well were incubated with 10 μL of CCK-8 reagent (Dojindo, Kumamoto, Japan) for 4 h. Afterwards, the value of optical density at a wavelength of 450 nm was quantified using a microplate reader (Bio-Rad, Hercules, CA, USA). The experiment was repeated three times.
5-Ethynyl-2'-deoxyuridine (EdU) staining. EdU staining for cell proliferation was conducted using the EdU kit (Beyotime, Beijing, China). In brief, HANK1 cells were seeded into the 24-well plate (2 × 104 cells/well). Then, HANK1 cells were incubated with the EdU reagent for 3 h, fixed with 4% paraformaldehyde for 15 min, and treated with 0.3% Triton X-100 for 15 min. After that, HANK1 cells were incubated with the Click Reaction Mixture in the dark at room temperature for 30 min and then treated with 4',6-diamidino-2-phenylindole for another 30 min. Afterwards, cells were observed under a fluorescence microscope (Olympus, Tokyo, Japan) and the results were displayed as a percentage. The experiment was repeated three times.
Co-immunoprecipitation (Co-IP) assay. In the Co-IP assay, HANK1 cells were lysed in the lysis buffer consisting of 150 mmol/L NaCl, 50 mmol/L Tris-HCl (pH = 7.6), 1% NP-40, 1 mmol/L ethylene diamine tetraacetic acid, 1 mmol/L NaF, 1 mmol/L Na3VO4, 1 mmol/L phenylmethanesulfonylfluoride, and 0.1% protease inhibitor cocktail. After centrifugation, the supernatant was collected and incubated with the antibody against TRAF6 (ab137452, Abcam, Cambridge, MA, USA) or IgG (ab133470, Abcam) at 4℃ for 30 min. Then, the supernatant was blended with protein A/G Plus-Agarose beads (Santa Cruz Biotechnology, Santa Cruz, MA, USA) using a shaker at 4℃ overnight. After three washes, the lysates were boiled in sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) protein buffer, followed by determination of the MST1 protein level using Western blot analysis. The experiment was repeated three times.
Ubiquitination assay. HANK1 cells were treated with 20 μM MG132. After 4 h, cells were collected and lysed using the lysis buffer. Then, lysates were incubated with the antibody against MST1 (ab264127, Abcam) or IgG (ab133470, Abcam) and then incubated with protein A/G plus agarose beads at 4℃ overnight. Next, the ubiquitination level of MST1 (ab19247, Abcam) was determined by Western blot analysis. The experiment was repeated three times.
Real-time quantitative polymerase chain reaction (RT-qPCR). According to the instructions provided by Invitrogen, the total RNA was extracted from cultured cell lines using the TRIzol reagent. Then, 1 μg of total RNA was reverse transcribed into the complementary DNA using the PrimeScript RT Reagent kit (Invitrogen). RT-qPCR was performed using SYBR green real-time Master Mix (TaKaRa, Dalian, China) on the 7500 Sequence Detection system (Applied Biosystems, Foster City, CA, USA). The relative gene expression was normalized to GAPDH and calculated using the 2-ΔΔCt method.[25] Primers used in PCR are shown in Table 1. The experiment was repeated three times.
Western blot analysis. Cultured cell lines were lysed in the radioimmunoprecipitation assay buffer (Beyotime), and lysates were ultrasonically processed, cultured on ice for 30 min, and centrifuged at 4℃ and 12000 g for 15 min. The protein density was quantified using the bicinchoninic acid kit (Beyotime). Protein samples were isolated using 8% SDS-PAGE, transferred onto nitrocellulose membranes, and immediately blocked with 2% bovine serum albumin for one hour. After that, membranes were incubated at 4℃ overnight with antibodies against proliferating cell nuclear antigen (PCNA) (ab92552, 1:1000), TRAF6 (ab137452, 1:500), MST1 (ab264127, 1:500), and β-actin (ab8227, 1:1000) and then incubated with the secondary antibody (1:2000, ab205718) for 1 h. Western blots were visualized using the enhanced chemiluminescence SuperSignal reagent (Thermo Fisher Scientific, Waltham, MA, USA). The grayscale value of target bands was analyzed using ImageJ software. All antibodies were provided by Abcam. The representative original bands are provided in the Supplementary file. The experiment was repeated three times.
Statistical analysis. Statistical analysis and graphing of data were performed with SPSS21.0 statistical software (IBM Corp, Armonk, NY, USA) and GraphPad Prism 8.0 software (GraphPad Software Inc., San Diego, CA, USA). Data complied with normal distribution and homogeneity of variance. The t-test was performed for pairwise comparisons of data, and one-way analysis of variance (ANOVA) was performed for multi-group comparisons of data, followed by Tukey's multiple comparison test for the post-test of data. The value of P < 0.05 referred to statistical significance.
Results
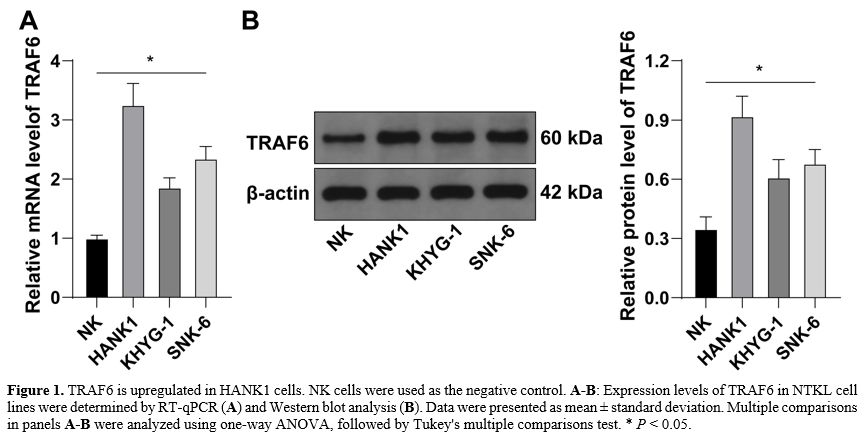
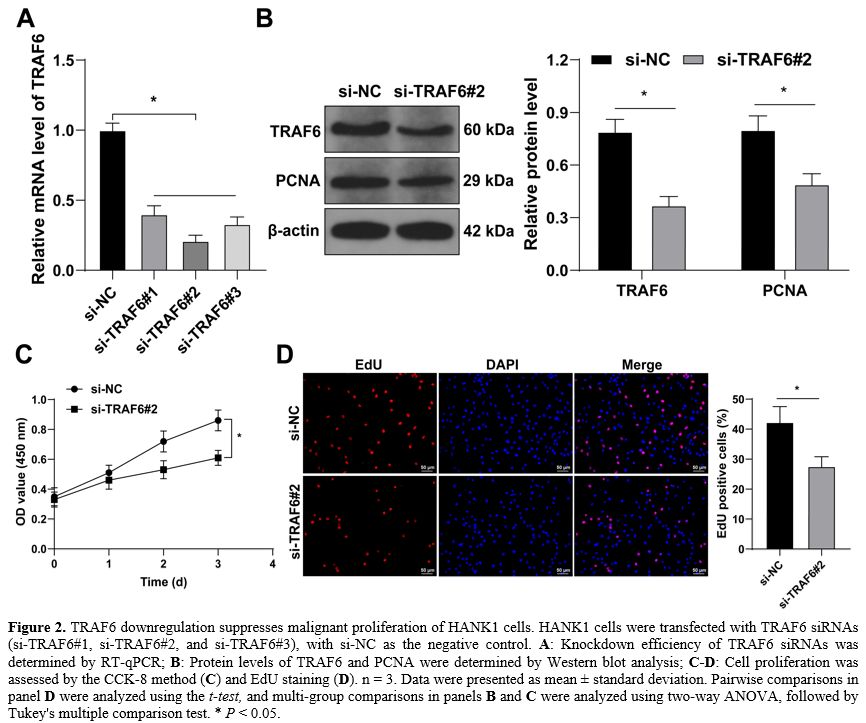
TRAF6 is upregulated in HANK1 cells. TRAF6 has been reported to be expressed at high levels in NKTL cells.[14] However, its role in HANK1 cell proliferation remains elusive. Firstly, we determined TRAF6 expression levels in NTKL cell lines and found that TRAF6 was upregulated in NTKL cell lines relative to normal NK cells, with HANK1 cells showing significantly increased expression of TRAF6 (Figure 1A-B).TRAF6 downregulation suppresses the malignant proliferation of HANK1 cells. To explore the impact of TRAF6 on HANK1 cell proliferation, HANK1 cells were transfected with TRAF6 siRNAs (si-TRAF6#1, si-TRAF6#2, and si-TRAF6#3) to downregulate TRAF6 expression in cells (P < 0.05, Figure 2A-B). si-TRAF6#2 was found to have the highest knockdown efficiency, and as a result, was selected for the subsequent experiment. The ensuing experiments further showed that after knockdown of TRAF6, the protein levels of proliferation marker PCNA were decreased (P < 0.05, Figure 2B), cell potential for proliferation was weakened (P < 0.05, Figure 2C). The number of EdU-positive cells was reduced (P < 0.05, Figure 2D). These findings suggested that TRAF6 downregulation suppresses malignant proliferation of HANK1 cells.
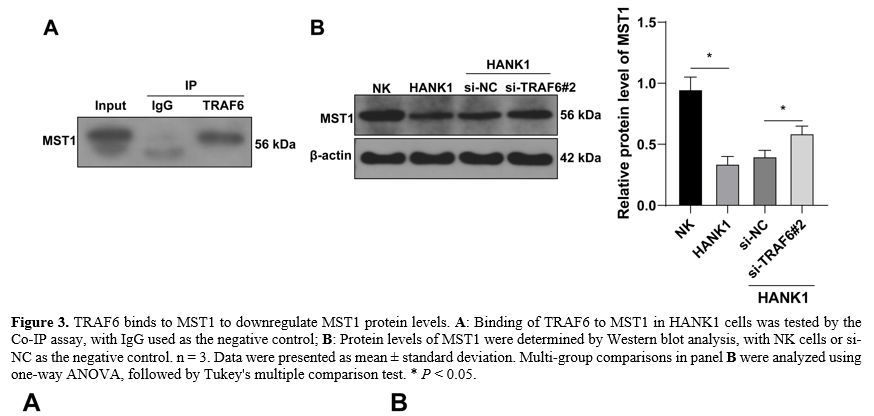
TRAF6 binds to MST1 to downregulate MST1 protein levels. It has been demonstrated that TRAF6 binds to MST1 to promote the ubiquitination-mediated degradation of MST1,[23] and MST1 is weakly expressed in NKTL [24] Therefore, the Co-IP assay was performed in HANK1 cells. The findings revealed the binding of TRAF6 to MST1 (Figure 3A). Findings of Western blot assay showed that the protein levels of MST1 were downregulated in HANK1 cells compared with normal NK cells and were increased after knockdown of TRAF6 (P < 0.05, Figure 3B). Above all, these findings suggested that TRAF6 bound to MST1 to downregulate MST1 protein levels.
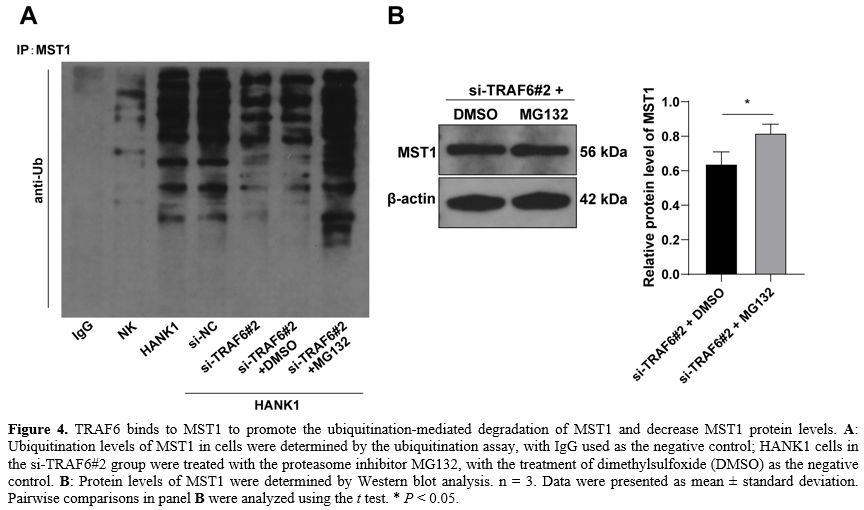
TRAF6 binds to MST1 to promote the ubiquitination-mediated degradation of MST1 and decrease MST1 protein levels. To confirm whether TRAF6 regulates MST1 via ubiquitination modification, the ubiquitination levels of MST1 in cells were determined and found to be markedly elevated in HANK1 cells and decreased as a result of the knockdown of TRAF6 (Figure 4A). Next, HANK1 cells were treated with the proteasome inhibitor MG132 to accumulate the ubiquitination levels of MST1 in cells (Figure 4A), upon which the protein levels of MST1 were further augmented (P < 0.05, Figure 4B). These findings suggested that TRAF6 binds to MST1 to promote the ubiquitination-mediated degradation of MST1 and decrease MST1 protein levels.
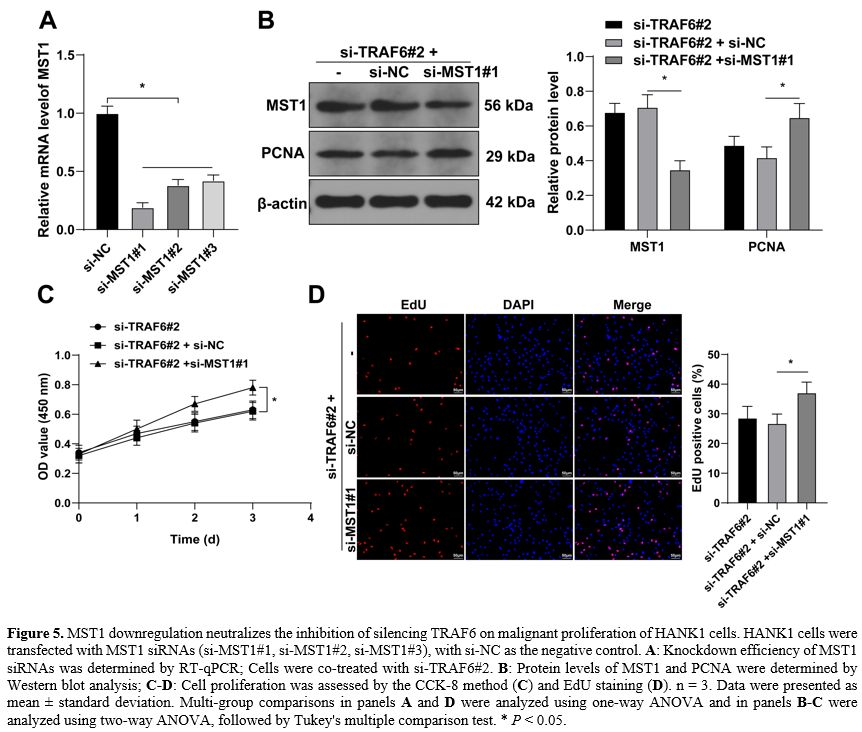
MST1 downregulation neutralizes the inhibition of silencing TRAF6 on the malignant proliferation of HANK1 cells. At last, HANK1 cells were transfected with MST1 siRNAs (si-MST1#1, si-MST1#2, si-MST1#3) to downregulate MST1 expression (P < 0.05, Figure 5A-B), and si-MST1#1 was found to have the highest knockdown efficiency and was selected for combined treatment with si-TRAF6#2. Our results showed that after knockdown of MST1, protein levels of PCNA increased (P < 0.05, Figure 5B), cell potential for proliferation was strengthened (P < 0.05, Figure 5C), and the number of EdU-positive cells was augmented (P < 0.05, Figure 5D). These findings suggested that MST1 downregulation neutralized the inhibition of silencing TRAF6 on malignant proliferation of HANK1 cells.
Discussion
NKTL is a highly aggressive malignancy, with few therapies effective in prolonging the survival time of affected patients.[26] Exploring epigenetic aberrations in NKTL helps identify effective targets for the treatment of NKTL.[26] TRAF6 has been demonstrated to play a role in the epigenetic regulation in various cancers.[10,11] However, relevant data on the molecular mechanism of TRAF6 in NKTL are largely limited. In the current study, our findings suggested that TRAF6 promoted the ubiquitination-mediated degradation of MST1 to facilitate malignant proliferation of HANK1 cells. Our study is the first to report the role of the TRAF6/MST1 axis in HANK1 cells, providing a new theoretical basis for the diagnosis and treatment of NKTL.TRAF6 has been identified as a positive regulator of lymphoma biology.[16] As an adaptor molecule, TRAF6 plays a prominent role in signal transduction of the NF-κB signaling, leading to lymphomagenesis.[27] Notably, the TRAF6/NF-κB pathway forms a negative feedback loop with miR-146a to regulate apoptosis and chemosensitivity in NKTL.[14] PCNA is a well-documented factor that is indicative of cancer cell proliferation.[28] In our experiments, TRAF6 was upregulated in HANK1 cells, and silencing TRAF6 repressed the malignant proliferation of HANK1 cells along with decreased protein levels of PCNA. Consistently, former studies have elucidated that TRAF6 interacts with LIM domain-containing protein 1 to prevent DNA damage and apoptosis and promote autophagy in EBV-associated lymphomas[29,30] and TRAF6 is involved in lncRNA NORAD-mediated oncogenic competitive endogenous RNA network to promote cell proliferation and inhibit cell cycle arrest and apoptosis in diffuse large B cell lymphoma (DLBCL).[17] Besides, inhibition of TRAF6 displays therapeutic implications in lymphomas. For instance, TRAF6 depletion caused cell cycle arrest at the G0/G1 phase and cell death of DLBCL.[31] High expression of TRAF6 correlates with poor prognosis and silencing of TRAF6 increases the sensitivity of DLBCL cells to doxorubicin.[16] Collectively, our findings and existing evidence both support the prominent expression of TRAF6 in HANK1 cells and its promotive role in the malignant proliferation of HANK1 cells.
With Lys of MST1 as a site of TRAF6-mediated ubiquitination,[32] TRAF6 has been reported to catalyze the ubiquitination of MST1 to repress its expression.[23] A pioneering study has highlighted the correlation between the MST1 mutation and the onset of NKTL.[33] Another study has also reported the downregulation of MST1 in NKTL tissues and the promotive role of MST1 knockdown in NKTL cell growth.[24] Besides, MST1 deficiency facilitates lymphoma development by causing chromosomal instability.[34] In accordance, our experiments uncovered reduced protein levels and increased ubiquitination levels of MST1 in HANK1 cells, and these trends were reversed by TRAF6 downregulation or MG132 treatment, suggesting that TRAF6 bound to MST1 to promote the ubiquitination-mediated degradation of MST1 and decrease MST1 protein levels in HANK1 cells. Furthermore, silencing MST1 decreased protein levels of PCNA and enhanced malignant proliferation of HANK1 cells in the si-TRAF6 group. Supporting our findings, Hippo pathways, including MST1, are found to be associated with the progression-free survival of B-cell lymphoma.[35] Moreover, as a tumor suppressor in the Hippo pathways, MST1 can be independent of or involved in the Hippo signaling pathway to inhibit the progression of many cancer types.[36,37] For instance, MST1 triggers reactive oxygen species-mediated pyroptosis to slow down the progression of pancreatic cancer;[38] MST1 activates the Hippo signaling pathway to enhance the malignant behaviors of breast cancer cells.[39] Above all, we are the first to unveil that TRAF6 promoted malignant proliferation of HANK1 cells via inducing ubiquitination of MST1, which underpinned the novelty of our study.
Conclusions
Our study initially demonstrated the connection of the TRAF6/MST1 signaling in malignant proliferation of HANK1 cells and provided a novel insight into the application of targeted therapy for NKTL treatment. The aberrant activation of TRAF6 in NKTL may serve as a molecular feature distinguishing it from other lymphoma subtypes. The TRAF6/MST1 signaling may target specific other signaling networks (including MST1) in the tumor microenvironment, thereby addressing the gap in resistance to existing therapies (e.g., PD-1 inhibitors) and exhibiting potential synergistic effects, especially in relapsed/refractory cases. In future studies, we will supplement data on long-term survival rates and evaluate the impact of targeted therapy on immune reconstitution. However, our current investigation of TRAF6 functionality and downstream mechanism was limited to HANK1 cells; the roles of TRAF6 in other cellular processes of NKTL cells, such as invasion and migration, remain unknown; the role of TRAF6 in NKTL was not validated in animal models; the function of complete TRAF6 knockout on NKTL cells requires further verification; phosphorylation plays an important role in ubiquitination modification, however, the effect of TRAF6 on MST1 phosphorylation cannot be verified under our current conditions; other downstream mechanisms of TRAF6 in NKTL, except MST1, remain to be explored. In the next step, we shall validate other mechanisms of TRAF6 using more NKTL cell lines, employ various experimental approaches to explore the novel downstream mechanisms of TRAF6, conduct preliminary experiments to verify the effect of TRAF6 on MST1 phosphorylation, and comprehensively investigate the impact of TRAF6 on the malignant phenotype of NKTL, providing updated theoretical knowledge for NKTL treatment.Acknowledgements
This work was supported by the Joint Funds for the innovation of science and Technology, Fujian province (Grant number: 2024Y9495). Fujian provincial health technology project (Grant number: 2022QNA102). Fujian Provincial Natural Science Foundation of China (Grant number: 2025J01899).References
- He X, Gao Y, Li Z, Huang H. Review on natural killer/T-cell lymphoma. Hematol Oncol. 2023 Apr;41(2):221-229. https://doi.org/10.1002/hon.2944 PMid:34731509
- Xue W, Zhang M. Updating targets for natural killer/T-cell lymphoma immunotherapy. Cancer Biol Med. 2021 Feb 15;18(1):52-62. https://doi.org/10.20892/j.issn.2095-3941.2020.0400 PMid:33628584 PMCid:PMC7877170
- Wang H, Fu BB, Gale RP, Liang Y. NK-/T-cell lymphomas. Leukemia. 2021 Sep;35(9):2460-2468. https://doi.org/10.1038/s41375-021-01313-2 PMid:34117356 PMCid:PMC8410593
- Tse E, Kwong YL. The diagnosis and management of NK/T-cell lymphomas. J Hematol Oncol. 2017 Apr 14;10(1):85. https://doi.org/10.1186/s13045-017-0452-9 PMid:28410601 PMCid:PMC5391564
- Tralongo
P, Bakacs A, Larocca LM. EBV-Related Lymphoproliferative Diseases: A
Review in Light of New Classifications. Mediterr J Hematol Infect Dis.
2024 16(1):e2024042. https://doi.org/10.4084/MJHID.2024.042 PMid:38882456 PMCid:PMC11178045
- Xiong
J, Cui BW, Wang N, Dai YT, Zhang H, Wang CF, Zhong HJ, Cheng S, Ou-Yang
BS, Hu Y, Zhang X, Xu B, Qian WB, Tao R, Yan F, Hu JD, Hou M, Ma XJ,
Wang X, Liu YH, Zhu ZM, Huang XB, Liu L, Wu CY, Huang L, Shen YF, Huang
RB, Xu JY, Wang C, Wu DP, Yu L, Li JF, Xu PP, Wang L, Huang JY, Chen
SJ, Zhao WL. Genomic and Transcriptomic Characterization of Natural
Killer T Cell Lymphoma. Cancer Cell. 2020 Mar 16;37(3):403-419 e406.https://doi.og/10.1016/j.ccell.2020.02.005 PMid:32183952
- Allen PB, Lechowicz MJ. Management of NK/T-Cell Lymphoma, Nasal Type. J Oncol Pract. 2019 Oct;15(10):513-520. https://doi.org/10.1200/JOP.18.00719 PMid:31600461 PMCid:PMC6790879
- Shi
JH, Sun SC. Tumor Necrosis Factor Receptor-Associated Factor Regulation
of Nuclear Factor kappaB and Mitogen-Activated Protein Kinase Pathways.
Front Immunol. 2018 9(1849. https://doi.org/10.3389/fimmu.2018.01849 PMid:30140268 PMCid:PMC6094638
- Masperone
L, Codrich M, Persichetti F, Gustincich S, Zucchelli S, Legname G. The
E3 Ubiquitin Ligase TRAF6 Interacts with the Cellular Prion Protein and
Modulates Its Solubility and Recruitment to Cytoplasmic
p62/SQSTM1-Positive Aggresome-Like Structures. Mol Neurobiol. 2022
Mar;59(3):1577-1588. https://doi.org/10.1007/s12035-021-02666-6 PMid:35000151
- Wang
J, Wu X, Jiang M, Tai G. Mechanism by which TRAF6 Participates in the
Immune Regulation of Autoimmune Diseases and Cancer. Biomed Res Int.
2020 2020(4607197. https://doi.org/10.1155/2020/4607197 PMid:33294443 PMCid:PMC7714562
- Li J, Liu N, Tang L, Yan B, Chen X, Zhang J, Peng C. The relationship between TRAF6 and tumors. Cancer Cell Int. 2020 20(429. https://doi.org/10.1186/s12935-020-01517-z PMid:32905356 PMCid:PMC7469280
- Wang
H, Zhang Y, Wu X, Wang Y, Cui H, Li X, Zhang J, Tun N, Peng Y, Yu J.
Regulation of Human Natural Killer Cell IFN-gamma Production by
MicroRNA-146a via Targeting the NF-kappaB Signaling Pathway. Front
Immunol. 2018 9(293. https://doi.org/10.3389/fimmu.2018.00293 PMid:29593706 PMCid:PMC5854688
- Lan
P, Fan Y, Zhao Y, Lou X, Monsour HP, Zhang X, Choi Y, Dou Y, Ishii N,
Ghobrial RM, Xiao X, Li XC. TNF superfamily receptor OX40 triggers
invariant NKT cell pyroptosis and liver injury. J Clin Invest. 2017 Jun
1;127(6):2222-2234. https://doi.org/10.1172/JCI91075 PMid:28436935 PMCid:PMC5451219
- Paik
JH, Jang JY, Jeon YK, Kim WY, Kim TM, Heo DS, Kim CW. MicroRNA-146a
downregulates NFkappaB activity via targeting TRAF6 and functions as a
tumor suppressor having strong prognostic implications in NK/T cell
lymphoma. Clin Cancer Res. 2011 Jul 15;17(14):4761-4771. https://doi.org/10.1158/1078-0432.CCR-11-0494 PMid:21610143
- Wu
CH, Yang YH, Chen MR, Tsai CH, Cheng AL, Doong SL. Autocleavage of the
paracaspase MALT1 at Arg-781 attenuates NF-kappaB signaling and
regulates the growth of activated B-cell like diffuse large B-cell
lymphoma cells. PLoS One. 2018 13(6):e0199779. https://doi.org/10.1371/journal.pone.0199779 PMid:29953499 PMCid:PMC6023146
- Qu
C, Kunkalla K, Vaghefi A, Frederiksen JK, Liu Y, Chapman JR, Blonska M,
Bernal-Mizrachi L, Alderuccio JP, Lossos IS, Landgraf R, Vega F.
Smoothened stabilizes and protects TRAF6 from degradation: A novel
non-canonical role of smoothened with implications in lymphoma biology.
Cancer Lett. 2018 Nov 1;436(149-158. https://doi.org/10.1016/j.canlet.2018.08.020 PMid:30165192
- Li
Y, Lv Y, Wang J, Zhu X, Chen J, Zhang W, Wang C, Jiang L. LncRNA NORAD
Mediates the Proliferation and Apoptosis of Diffuse Large-B-Cell
Lymphoma via Regulation of miR-345-3p/TRAF6 Axis. Arch Med Res. 2022
Apr;53(3):271-279. https://doi.org/10.1016/j.arcmed.2022.01.004 PMid:35164979
- Mansour MA. Ubiquitination: Friend and foe in cancer. Int J Biochem Cell Biol. 2018 Aug;101(80-93. https://doi.org/10.1016/j.biocel.2018.06.001 PMid:29864543
- Wu
H, Lu XX, Wang JR, Yang TY, Li XM, He XS, Li Y, Ye WL, Wu Y, Gan WJ,
Guo PD, Li JM. TRAF6 inhibits colorectal cancer metastasis through
regulating selective autophagic CTNNB1/beta-catenin degradation and is
targeted for GSK3B/GSK3beta-mediated phosphorylation and degradation.
Autophagy. 2019 Sep;15(9):1506-1522. https://doi.org/10.1080/15548627.2019.1586250 PMid:30806153 PMCid:PMC6693460
- Wen
F, Sun X, Sun C, Dong Z, Jia G, Bao W, Yu H, Yang C. TAGLN Is
Downregulated by TRAF6-Mediated Proteasomal Degradation in Prostate
Cancer Cells. Mol Cancer Res. 2021 Jul;19(7):1113-1122. https://doi.org/10.1158/1541-7786.MCR-20-0513 PMid:33771884
- Song
G, Zhang Y, Tian J, Ma J, Yin K, Xu H, Wang S. TRAF6 Regulates the
Immunosuppressive Effects of Myeloid-Derived Suppressor Cells in
Tumor-Bearing Host. Front Immunol. 2021 12(649020. https://doi.org/10.3389/fimmu.2021.649020 PMid:33717204 PMCid:PMC7946975
- Taha Z, Janse van Rensburg HJ, Yang X. The Hippo Pathway: Immunity and Cancer. Cancers (Basel). 2018 Mar 28;10(4) https://doi.org/10.3390/cancers10040094 PMid:29597279 PMCid:PMC5923349
- Li
JA, Kuang T, Pu N, Fang Y, Han X, Zhang L, Xu X, Wu W, Wang D, Lou W,
Rong Y. TRAF6 regulates YAP signaling by promoting the ubiquitination
and degradation of MST1 in pancreatic cancer. Clin Exp Med. 2019
May;19(2):211-218. https://doi.org/10.1007/s10238-018-00543-6 PMid:30673917
- Chang
Y, Fu XR, Cui M, Li WM, Zhang L, Li X, Li L, Sun ZC, Zhang XD, Li ZM,
You XY, Nan FF, Wu JJ, Wang XH, Zhang MZ. Activated hippo signal
pathway inhibits cell proliferation and promotes apoptosis in NK/T cell
lymphoma cells. Cancer Med. 2019 Jul;8(8):3892-3904. https://doi.org/10.1002/cam4.2174 PMid:31124291 PMCid:PMC6639190
- Livak
KJ, Schmittgen TD. Analysis of relative gene expression data using
real-time quantitative PCR and the 2(-Delta Delta C(T)) Method.
Methods. 2001 Dec;25(4):402-408. https://doi.org/10.1006/meth.2001.1262 PMid:11846609
- Kucuk
C, Wang J, Xiang Y, You H. Epigenetic aberrations in natural
killer/T-cell lymphoma: diagnostic, prognostic and therapeutic
implications. Ther Adv Med Oncol. 2020 12(1758835919900856. https://doi.org/10.1177/1758835919900856 PMid:32127923 PMCid:PMC7036507
- Rosebeck
S, Lim MS, Elenitoba-Johnson KS, McAllister-Lucas LM, Lucas PC.
API2-MALT1 oncoprotein promotes lymphomagenesis via unique program of
substrate ubiquitination and proteolysis. World J Biol Chem. 2016 Feb
26;7(1):128-137. https://doi.org/10.4331/wjbc.v7.i1.128 PMid:26981201 PMCid:PMC4768116
- Cardano
M, Tribioli C, Prosperi E. Targeting Proliferating Cell Nuclear Antigen
(PCNA) as an Effective Strategy to Inhibit Tumor Cell Proliferation.
Curr Cancer Drug Targets. 2020 20(4):240-252. https://doi.org/10.2174/1568009620666200115162814 PMid:31951183
- Wang
L, Howell MEA, Sparks-Wallace A, Zhao J, Hensley CR, Nicksic CA, Horne
SR, Mohr KB, Moorman JP, Yao ZQ, Ning S. The Ubiquitin Sensor and
Adaptor Protein p62 Mediates Signal Transduction of a Viral Oncogenic
Pathway. mBio. 2021 Oct 26;12(5):e0109721. https://doi.org/10.1128/mBio.01097-21 PMid:34488443 PMCid:PMC8546576
- Wang
L, Howell MEA, McPeak B, Riggs K, Kohne C, Yohanon JU, Foxler DE, Sharp
TV, Moorman JP, Yao ZQ, Ning S. LIMD1 is induced by and required for
LMP1 signaling, and protects EBV-transformed cells from DNA
damage-induced cell death. Oncotarget. 2018 Jan 19;9(5):6282-6297. https://doi.org/10.18632/oncotarget.23676 PMid:29464072 PMCid:PMC5814212
- Dai
YH, Hung LY, Chen RY, Lai CH, Chang KC. ON 01910.Na inhibits growth of
diffuse large B-cell lymphoma by cytoplasmic sequestration of
sumoylated C-MYB/TRAF6 complex. Transl Res. 2016 Sep;175(129-143 e113. https://doi.org/10.1016/j.trsl.2016.04.001 PMid:27150054
- Roh
KH, Lee Y, Yoon JH, Lee D, Kim E, Park E, Lee IY, Kim TS, Song HK, Shin
J, Lim DS, Choi EJ. TRAF6-mediated ubiquitination of MST1/STK4
attenuates the TLR4-NF-kappaB signaling pathway in macrophages. Cell
Mol Life Sci. 2021 Mar;78(5):2315-2328. https://doi.org/10.1007/s00018-020-03650-4 PMid:32975614 PMCid:PMC11071754
- Wang
GN, Zhao WG, Zhang XD, Jian XY, Zhang CL, Zhang MZ, Li WC. A
retrospective study on the clinicopathological and molecular features
of 22 cases of natural killer/T-cell lymphoma in children and
adolescents. Sci Rep. 2022 May 3;12(1):7118. https://doi.org/10.1038/s41598-022-11247-z PMid:35504960 PMCid:PMC9064969
- Kim
TS, Lee DH, Kim SK, Shin SY, Seo EJ, Lim DS. Mammalian sterile 20-like
kinase 1 suppresses lymphoma development by promoting faithful
chromosome segregation. Cancer Res. 2012 Oct 15;72(20):5386-5395. https://doi.org/10.1158/0008-5472.CAN-11-3956 PMid:22926556
- Fu
H, Zhou H, Qiu Y, Wang J, Ma Z, Li H, Zhang F, Qiu C, Shen J, Liu T.
SEPT6_TRIM33 Gene Fusion and Mutated TP53 Pathway Associate With
Unfavorable Prognosis in Patients With B-Cell Lymphomas. Front Oncol.
2021 11(765544. https://doi.org/10.3389/fonc.2021.765544 PMid:34926267 PMCid:PMC8671703
- Wang
Y, Jia A, Cao Y, Hu X, Wang Y, Yang Q, Bi Y, Liu G. Hippo Kinases
MST1/2 Regulate Immune Cell Functions in Cancer, Infection, and
Autoimmune Diseases. Crit Rev Eukaryot Gene Expr. 2020 30(5):427-442. https://doi.org/10.1615/CritRevEukaryotGeneExpr.2020035775 PMid:33389879
- Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010 Oct 19;19(4):491-505. https://doi.org/10.1016/j.devcel.2010.09.011 PMid:20951342 PMCid:PMC3124840
- Cui
J, Zhou Z, Yang H, Jiao F, Li N, Gao Y, Wang L, Chen J, Quan M. MST1
Suppresses Pancreatic Cancer Progression via ROS-Induced Pyroptosis.
Mol Cancer Res. 2019 Jun;17(6):1316-1325. https://doi.org/10.1158/1541-7786.MCR-18-0910 PMid:30796177
- Jin
X, Zhu L, Xiao S, Cui Z, Tang J, Yu J, Xie M. MST1 inhibits the
progression of breast cancer by regulating the Hippo signaling pathway
and may serve as a prognostic biomarker. Mol Med Rep. 2021 May;23(5) https://doi.org/10.3892/mmr.2021.12022 PMid:33760220 PMCid:PMC7986037